The birth of the Epitranscriptome: deciphering the function of RNA modifications
- PMID: 23113984
- PMCID: PMC3491402
- DOI: 10.1186/gb-2012-13-10-175
The birth of the Epitranscriptome: deciphering the function of RNA modifications
Abstract
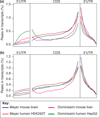
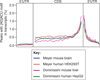
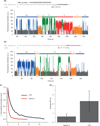
Recent studies have found methyl-6-adenosine in thousands of mammalian genes, and this modification is most pronounced near the beginning of the 3' UTR. We present a perspective on current work and new single-molecule sequencing methods for detecting RNA base modifications.
Figures

Similar articles
-
m(6)A-LAIC-seq reveals the census and complexity of the m(6)A epitranscriptome.Nat Methods. 2016 Aug;13(8):692-8. doi: 10.1038/nmeth.3898. Epub 2016 Jul 4. Nat Methods. 2016. PMID: 27376769 Free PMC article.
-
Novel RNA regulatory mechanisms revealed in the epitranscriptome.RNA Biol. 2013 Mar;10(3):342-6. doi: 10.4161/rna.23812. Epub 2013 Feb 22. RNA Biol. 2013. PMID: 23434792 Free PMC article. Review.
-
Detecting RNA modifications in the epitranscriptome: predict and validate.Nat Rev Genet. 2017 May;18(5):275-291. doi: 10.1038/nrg.2016.169. Epub 2017 Feb 20. Nat Rev Genet. 2017. PMID: 28216634 Review.
-
Deciphering the epitranscriptome: A green perspective.J Integr Plant Biol. 2016 Oct;58(10):822-835. doi: 10.1111/jipb.12483. Epub 2016 Jun 20. J Integr Plant Biol. 2016. PMID: 27172004 Free PMC article. Review.
-
Misincorporation signatures for detecting modifications in mRNA: Not as simple as it sounds.Methods. 2019 Mar 1;156:53-59. doi: 10.1016/j.ymeth.2018.10.011. Epub 2018 Oct 23. Methods. 2019. PMID: 30359724 Review.
Cited by
-
The advantages of SMRT sequencing.Genome Biol. 2013 Jul 3;14(7):405. doi: 10.1186/gb-2013-14-6-405. Genome Biol. 2013. PMID: 23822731 Free PMC article.
-
Characterizing Post-transcriptional Modifications of circRNAs to Investigate Biogenesis and Translation.Methods Mol Biol. 2024;2765:263-296. doi: 10.1007/978-1-0716-3678-7_15. Methods Mol Biol. 2024. PMID: 38381345
-
Up-regulation of RNA m6A methyltransferase like-3 expression contributes to arsenic and benzo[a]pyrene co-exposure-induced cancer stem cell-like property and tumorigenesis.Toxicol Appl Pharmacol. 2023 Dec 15;481:116764. doi: 10.1016/j.taap.2023.116764. Epub 2023 Nov 14. Toxicol Appl Pharmacol. 2023. PMID: 37972769 Free PMC article.
-
Next-generation sequencing: a frameshift in skeletal dysplasia gene discovery.Osteoporos Int. 2014 Feb;25(2):407-22. doi: 10.1007/s00198-013-2443-1. Epub 2013 Aug 1. Osteoporos Int. 2014. PMID: 23903953 Review.
-
YTHDF2 Recognition of N1-Methyladenosine (m1A)-Modified RNA Is Associated with Transcript Destabilization.ACS Chem Biol. 2020 Jan 17;15(1):132-139. doi: 10.1021/acschembio.9b00655. Epub 2019 Dec 12. ACS Chem Biol. 2020. PMID: 31815430 Free PMC article.
References
-
- Johnson TB, Coghill RD. Researches on pyrimidines. C111. The discovery of 5-methyl-cytosine in tuberculnic acid, the nucleic acid of the tubercle bacillis. J Am Chem Soc. 1925;47:2838–2844. doi: 10.1021/ja01688a030. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

