Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells
- PMID: 23101633
- PMCID: PMC3482660
- DOI: 10.1016/j.cell.2012.09.019
Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells
Abstract
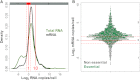
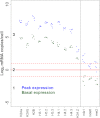
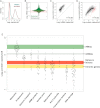
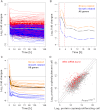
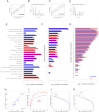
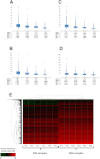
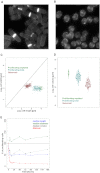
Data on absolute molecule numbers will empower the modeling, understanding, and comparison of cellular functions and biological systems. We quantified transcriptomes and proteomes in fission yeast during cellular proliferation and quiescence. This rich resource provides the first comprehensive reference for all RNA and most protein concentrations in a eukaryote under two key physiological conditions. The integrated data set supports quantitative biology and affords unique insights into cell regulation. Although mRNAs are typically expressed in a narrow range above 1 copy/cell, most long, noncoding RNAs, except for a distinct subset, are tightly repressed below 1 copy/cell. Cell-cycle-regulated transcription tunes mRNA numbers to phase-specific requirements but can also bring about more switch-like expression. Proteins greatly exceed mRNAs in abundance and dynamic range, and concentrations are regulated to functional demands. Upon transition to quiescence, the proteome changes substantially, but, in stark contrast to mRNAs, proteins do not uniformly decrease but scale with cell volume.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures





Comment in
-
Gene expression: dynamic omics responses.Nat Rev Genet. 2012 Dec;13(12):828. doi: 10.1038/nrg3370. Epub 2012 Nov 14. Nat Rev Genet. 2012. PMID: 23150041 No abstract available.
Similar articles
-
Absolute proteome and phosphoproteome dynamics during the cell cycle of Schizosaccharomyces pombe (Fission Yeast).Mol Cell Proteomics. 2014 Aug;13(8):1925-36. doi: 10.1074/mcp.M113.035824. Epub 2014 Apr 23. Mol Cell Proteomics. 2014. PMID: 24763107 Free PMC article.
-
Proteome analysis of Schizosaccharomyces pombe by two-dimensional gel electrophoresis and mass spectrometry.Proteomics. 2006 Jul;6(14):4115-29. doi: 10.1002/pmic.200500863. Proteomics. 2006. PMID: 16791824
-
Regulation of transcriptome, translation, and proteome in response to environmental stress in fission yeast.Genome Biol. 2012 Apr 18;13(4):R25. doi: 10.1186/gb-2012-13-4-r25. Genome Biol. 2012. PMID: 22512868 Free PMC article.
-
The selective elimination of messenger RNA underlies the mitosis-meiosis switch in fission yeast.Proc Jpn Acad Ser B Phys Biol Sci. 2010;86(8):788-97. doi: 10.2183/pjab.86.788. Proc Jpn Acad Ser B Phys Biol Sci. 2010. PMID: 20948174 Free PMC article. Review.
-
Meiosis. New roles for RNA in fission yeast.Curr Biol. 1995 Jan 1;5(1):4-6. doi: 10.1016/s0960-9822(95)00002-9. Curr Biol. 1995. PMID: 7697346 Review.
Cited by
-
Analysis of the SNARE Stx8 recycling reveals that the retromer-sorting motif has undergone evolutionary divergence.PLoS Genet. 2021 Mar 31;17(3):e1009463. doi: 10.1371/journal.pgen.1009463. eCollection 2021 Mar. PLoS Genet. 2021. PMID: 33788833 Free PMC article.
-
RNA interference is essential for cellular quiescence.Science. 2016 Nov 11;354(6313):aah5651. doi: 10.1126/science.aah5651. Epub 2016 Oct 13. Science. 2016. PMID: 27738016 Free PMC article.
-
Accounting for experimental noise reveals that mRNA levels, amplified by post-transcriptional processes, largely determine steady-state protein levels in yeast.PLoS Genet. 2015 May 7;11(5):e1005206. doi: 10.1371/journal.pgen.1005206. eCollection 2015 May. PLoS Genet. 2015. PMID: 25950722 Free PMC article.
-
Single mammalian cells compensate for differences in cellular volume and DNA copy number through independent global transcriptional mechanisms.Mol Cell. 2015 Apr 16;58(2):339-52. doi: 10.1016/j.molcel.2015.03.005. Epub 2015 Apr 9. Mol Cell. 2015. PMID: 25866248 Free PMC article.
-
Proportionality: a valid alternative to correlation for relative data.PLoS Comput Biol. 2015 Mar 16;11(3):e1004075. doi: 10.1371/journal.pcbi.1004075. eCollection 2015 Mar. PLoS Comput Biol. 2015. PMID: 25775355 Free PMC article.
References
Supplemental References
-
- Bähler, J., Wu, J.Q., Longtine, M.S., Shah, N.G., McKenzie, A., III, Steever, A.B., Wach, A., Philippsen, P., and Pringle, J.R. (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943–951. - PubMed
-
- Elias, J.E., and Gygi, S.P. (2007). Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

