Granzyme M targets host cell hnRNP K that is essential for human cytomegalovirus replication
- PMID: 23099853
- PMCID: PMC3569982
- DOI: 10.1038/cdd.2012.132
Granzyme M targets host cell hnRNP K that is essential for human cytomegalovirus replication
Abstract
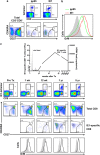
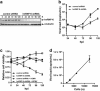
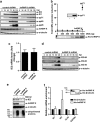
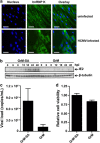
Human cytomegalovirus (HCMV) is the most frequent viral cause of congenital defects and HCMV infection in immunocompromised patients may trigger devastating disease. Cytotoxic lymphocytes control HCMV by releasing granzymes towards virus-infected cells. In mice, granzyme M (GrM) has a physiological role in controlling murine CMV infection. However, the underlying mechanism remains poorly understood. In this study, we showed that human GrM was expressed by HCMV-specific CD8(+) T cells both in latently infected healthy individuals and in transplant patients during primary HCMV infection. We identified host cell heterogeneous nuclear ribonucleoprotein K (hnRNP K) as a physiological GrM substrate. GrM most efficiently cleaved hnRNP K in the presence of RNA at multiple sites, thereby likely destroying hnRNP K function. Host cell hnRNP K was essential for HCMV replication not only by promoting viability of HCMV-infected cells but predominantly by regulating viral immediate-early 2 (IE2) protein levels. Furthermore, hnRNP K interacted with IE2 mRNA. Finally, GrM decreased IE2 protein expression in HCMV-infected cells. Our data suggest that targeting of hnRNP K by GrM contributes to the mechanism by which cytotoxic lymphocytes inhibit HCMV replication. This is the first evidence that cytotoxic lymphocytes target host cell proteins to control HCMV infections.
Figures


Similar articles
-
Killer cell proteases can target viral immediate-early proteins to control human cytomegalovirus infection in a noncytotoxic manner.PLoS Pathog. 2020 Apr 13;16(4):e1008426. doi: 10.1371/journal.ppat.1008426. eCollection 2020 Apr. PLoS Pathog. 2020. PMID: 32282833 Free PMC article.
-
Noncytotoxic inhibition of cytomegalovirus replication through NK cell protease granzyme M-mediated cleavage of viral phosphoprotein 71.J Immunol. 2010 Dec 15;185(12):7605-13. doi: 10.4049/jimmunol.1001503. Epub 2010 Nov 8. J Immunol. 2010. PMID: 21059895
-
Elevated granzyme M-expressing lymphocytes during cytomegalovirus latency and reactivation after allogeneic stem cell transplantation.Clin Immunol. 2014 Jan;150(1):1-11. doi: 10.1016/j.clim.2013.11.005. Epub 2013 Nov 18. Clin Immunol. 2014. PMID: 24316590
-
Bright and Early: Inhibiting Human Cytomegalovirus by Targeting Major Immediate-Early Gene Expression or Protein Function.Viruses. 2020 Jan 16;12(1):110. doi: 10.3390/v12010110. Viruses. 2020. PMID: 31963209 Free PMC article. Review.
-
Granzyme M: behind enemy lines.Cell Death Differ. 2014 Mar;21(3):359-68. doi: 10.1038/cdd.2013.189. Epub 2014 Jan 10. Cell Death Differ. 2014. PMID: 24413154 Free PMC article. Review.
Cited by
-
Cytomegalovirus-Infected Cells Resist T Cell Mediated Killing in an HLA-Recognition Independent Manner.Front Microbiol. 2016 Jun 9;7:844. doi: 10.3389/fmicb.2016.00844. eCollection 2016. Front Microbiol. 2016. PMID: 27375569 Free PMC article.
-
Perforin and granzymes: function, dysfunction and human pathology.Nat Rev Immunol. 2015 Jun;15(6):388-400. doi: 10.1038/nri3839. Nat Rev Immunol. 2015. PMID: 25998963 Review.
-
Identification of cellular proteins associated with human cytomegalovirus (HCMV) DNA replication suggests novel cellular and viral interactions.Virology. 2022 Jan;566:26-41. doi: 10.1016/j.virol.2021.11.004. Epub 2021 Nov 22. Virology. 2022. PMID: 34861458 Free PMC article.
-
Heterogeneous nuclear ribonucleoprotein K supports vesicular stomatitis virus replication by regulating cell survival and cellular gene expression.J Virol. 2013 Sep;87(18):10059-69. doi: 10.1128/JVI.01257-13. Epub 2013 Jul 10. J Virol. 2013. PMID: 23843646 Free PMC article.
-
Arginine methylation of hnRNPK negatively modulates apoptosis upon DNA damage through local regulation of phosphorylation.Nucleic Acids Res. 2014 Sep;42(15):9908-24. doi: 10.1093/nar/gku705. Epub 2014 Aug 7. Nucleic Acids Res. 2014. PMID: 25104022 Free PMC article.
References
-
- Mocarski ES, Shenk T, Pass RF.CytomegalovirusesIn: Knipe PM, Howley DE, Griffin DE, Lamb RA, Martin MA, Roizman B, et al. (eds)Fields Virology5th edn., Vol. 2Lippincott Williams & Wilkins: Philadelphia, PA; 20072701–2772.
-
- Moss P, Rickinson A. Cellular immunotherapy for viral infection after HSC transplantation. Nat Rev Immunol. 2005;5:9–20. - PubMed
-
- Gamadia LE, Remmerswaal EB, Weel JF, Bemelman F, van Lier RA, Ten Berge IJ. Primary immune responses to human CMV: a critical role for IFN-gamma-producing CD4+ T cells in protection against CMV disease. Blood. 2003;101:2686–2692. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

