HCMV gB shares structural and functional properties with gB proteins from other herpesviruses
- PMID: 23089254
- PMCID: PMC3534942
- DOI: 10.1016/j.virol.2012.09.024
HCMV gB shares structural and functional properties with gB proteins from other herpesviruses
Abstract
Glycoprotein B (gB) facilitates HCMV entry into cells by binding receptors and mediating membrane fusion. The crystal structures of gB ectodomains from HSV-1 and EBV are available, but little is known about the HCMV gB structure. Using multiangle light scattering and electron microscopy, we show here that HCMV gB ectodomain is a trimer with the overall shape similar to HSV-1 and EBV gB ectodomains. HCMV gB ectodomain forms rosettes similar to rosettes formed by EBV gB and the postfusion forms of other viral fusogens. Substitution of several bulky hydrophobic residues within the putative fusion loops with more hydrophilic residues reduced rosette formation and abolished cell fusion. We propose that like gB proteins from HSV-1 and EBV, HCMV gB has two internal hydrophobic fusion loops that likely interact with target membranes. Our work establishes structural and functional similarities between gB proteins from three subfamilies of herpesviruses.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures

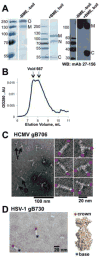

Similar articles
-
Structure-Function Dissection of Pseudorabies Virus Glycoprotein B Fusion Loops.J Virol. 2017 Dec 14;92(1):e01203-17. doi: 10.1128/JVI.01203-17. Print 2018 Jan 1. J Virol. 2017. PMID: 29046441 Free PMC article.
-
Fusion-deficient insertion mutants of herpes simplex virus type 1 glycoprotein B adopt the trimeric postfusion conformation.J Virol. 2010 Feb;84(4):2001-12. doi: 10.1128/JVI.01791-09. Epub 2009 Nov 25. J Virol. 2010. PMID: 19939928 Free PMC article.
-
The structural basis of herpesvirus entry.Nat Rev Microbiol. 2021 Feb;19(2):110-121. doi: 10.1038/s41579-020-00448-w. Epub 2020 Oct 21. Nat Rev Microbiol. 2021. PMID: 33087881 Free PMC article. Review.
-
The Fusion Loops of the Initial Prefusion Conformation of Herpes Simplex Virus 1 Fusion Protein Point Toward the Membrane.mBio. 2017 Aug 22;8(4):e01268-17. doi: 10.1128/mBio.01268-17. mBio. 2017. PMID: 28830949 Free PMC article.
-
Herpesvirus gB: A Finely Tuned Fusion Machine.Viruses. 2015 Dec 11;7(12):6552-69. doi: 10.3390/v7122957. Viruses. 2015. PMID: 26690469 Free PMC article. Review.
Cited by
-
Enveloped virus-like particle expression of human cytomegalovirus glycoprotein B antigen induces antibodies with potent and broad neutralizing activity.Clin Vaccine Immunol. 2014 Feb;21(2):174-80. doi: 10.1128/CVI.00662-13. Epub 2013 Dec 11. Clin Vaccine Immunol. 2014. PMID: 24334684 Free PMC article.
-
Enhanced expression of full-length human cytomegalovirus fusion protein in non-swelling baculovirus-infected cells with a minimal fed-batch strategy.PLoS One. 2014 Mar 4;9(3):e90753. doi: 10.1371/journal.pone.0090753. eCollection 2014. PLoS One. 2014. PMID: 24595278 Free PMC article.
-
Analysis of Cytomegalovirus Glycoprotein and Cellular Receptor Interactions.Methods Mol Biol. 2021;2244:199-211. doi: 10.1007/978-1-0716-1111-1_10. Methods Mol Biol. 2021. PMID: 33555588
-
Human Cytomegalovirus Congenital (cCMV) Infection Following Primary and Nonprimary Maternal Infection: Perspectives of Prevention through Vaccine Development.Vaccines (Basel). 2020 Apr 23;8(2):194. doi: 10.3390/vaccines8020194. Vaccines (Basel). 2020. PMID: 32340180 Free PMC article. Review.
-
Structural basis for the recognition of human cytomegalovirus glycoprotein B by a neutralizing human antibody.PLoS Pathog. 2014 Oct 9;10(10):e1004377. doi: 10.1371/journal.ppat.1004377. eCollection 2014 Oct. PLoS Pathog. 2014. PMID: 25299639 Free PMC article.
References
-
- Barton GJ. Alscript: a tool to format multiple sequence alignments. Protein Eng. 1993;6:37–40. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

