Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization
- PMID: 23086239
- PMCID: PMC3767400
- DOI: 10.1038/ncb2597
Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization
Abstract
As the main microtubule-organizing centre in animal cells, the centrosome has a fundamental role in cell function. Surrounding the centrioles, the pericentriolar material (PCM) provides a dynamic platform for nucleating microtubules. Although the importance of the PCM is established, its amorphous electron-dense nature has made it refractory to structural investigation. By using SIM and STORM subdiffraction-resolution microscopies to visualize proteins critical for centrosome maturation, we demonstrate that the PCM is organized into two main structural domains: a layer juxtaposed to the centriole wall, and proteins extending farther away from the centriole organized in a matrix. Analysis of Pericentrin-like protein (PLP) reveals that its carboxy terminus is positioned at the centriole wall, it radiates outwards into the matrix and is organized in clusters having quasi-nine-fold symmetry. By RNA-mediated interference (RNAi), we show that PLP fibrils are required for interphase recruitment and proper mitotic assembly of the PCM matrix.
Figures
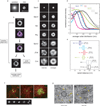
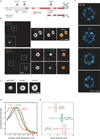

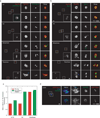
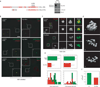


Comment in
-
Cell cycle: Order in the pericentriolar material.Nat Rev Mol Cell Biol. 2012 Dec;13(12):749. doi: 10.1038/nrm3471. Epub 2012 Oct 25. Nat Rev Mol Cell Biol. 2012. PMID: 23095798 No abstract available.
Similar articles
-
Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material.Nat Cell Biol. 2012 Nov;14(11):1148-58. doi: 10.1038/ncb2591. Epub 2012 Oct 21. Nat Cell Biol. 2012. PMID: 23086237
-
Self-assembly of pericentriolar material in interphase cells lacking centrioles.Elife. 2022 Jul 5;11:e77892. doi: 10.7554/eLife.77892. Elife. 2022. PMID: 35787744 Free PMC article.
-
Structured illumination of the interface between centriole and peri-centriolar material.Open Biol. 2012 Aug;2(8):120104. doi: 10.1098/rsob.120104. Open Biol. 2012. PMID: 22977736 Free PMC article.
-
Amorphous no more: subdiffraction view of the pericentriolar material architecture.Trends Cell Biol. 2014 Mar;24(3):188-97. doi: 10.1016/j.tcb.2013.10.001. Epub 2013 Nov 19. Trends Cell Biol. 2014. PMID: 24268653 Free PMC article. Review.
-
Human centrosome organization and function in interphase and mitosis.Semin Cell Dev Biol. 2021 Sep;117:30-41. doi: 10.1016/j.semcdb.2021.03.020. Epub 2021 Apr 6. Semin Cell Dev Biol. 2021. PMID: 33836946 Free PMC article. Review.
Cited by
-
SAPs as a new model to probe the pathway of centriole and centrosome assembly.Biochem Soc Trans. 2021 Jun 30;49(3):1233-1240. doi: 10.1042/BST20200833. Biochem Soc Trans. 2021. PMID: 33960367 Free PMC article. Review.
-
Quantitative analysis of human centrosome architecture by targeted proteomics and fluorescence imaging.EMBO J. 2016 Oct 4;35(19):2152-2166. doi: 10.15252/embj.201694462. Epub 2016 Aug 18. EMBO J. 2016. PMID: 27539480 Free PMC article.
-
Reflections on mentorship as an early career researcher.Mol Biol Cell. 2022 Dec 1;33(14):ae3. doi: 10.1091/mbc.E22-08-0359. Mol Biol Cell. 2022. PMID: 36399627 Free PMC article.
-
Evidence that a positive feedback loop drives centrosome maturation in fly embryos.Elife. 2019 Sep 9;8:e50130. doi: 10.7554/eLife.50130. Elife. 2019. PMID: 31498081 Free PMC article.
-
CEP215 and AURKA regulate spindle pole focusing and aMTOC organization in mouse oocytes.Reproduction. 2020 Mar;159(3):261-274. doi: 10.1530/REP-19-0263. Reproduction. 2020. PMID: 31895686 Free PMC article.
References
-
- Bettencourt-Dias M, Glover DM. Centrosome biogenesis and function: centrosomics brings new understanding. Nat Rev Mol Cell Biol. 2007;8(6):451–463. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

