Precision editing of large animal genomes
- PMID: 23084873
- PMCID: PMC3683964
- DOI: 10.1016/B978-0-12-404742-6.00002-8
Precision editing of large animal genomes
Abstract
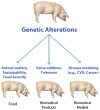
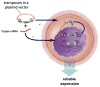
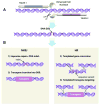
Transgenic animals are an important source of protein and nutrition for most humans and will play key roles in satisfying the increasing demand for food in an ever-increasing world population. The past decade has experienced a revolution in the development of methods that permit the introduction of specific alterations to complex genomes. This precision will enhance genome-based improvement of farm animals for food production. Precision genetics also will enhance the development of therapeutic biomaterials and models of human disease as resources for the development of advanced patient therapies.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Precision genetics for complex objectives in animal agriculture.J Anim Sci. 2010 Jul;88(7):2530-9. doi: 10.2527/jas.2010-2847. Epub 2010 Mar 12. J Anim Sci. 2010. PMID: 20228236 Free PMC article.
-
Transgenic farm animals: applications in agriculture and biomedicine.Biotechnol Annu Rev. 2000;5:269-92. doi: 10.1016/s1387-2656(00)05039-0. Biotechnol Annu Rev. 2000. PMID: 10875004 Review.
-
Genome editing for crop improvement: Challenges and opportunities.GM Crops Food. 2015;6(4):183-205. doi: 10.1080/21645698.2015.1129937. GM Crops Food. 2015. PMID: 26930114 Free PMC article.
-
How farm animals are improving human health and welfare.Rev Sci Tech. 2018 Apr;37(1):83-96. doi: 10.20506/rst.37.1.2742. Rev Sci Tech. 2018. PMID: 30209427 Review.
-
[Application of CRISPR/Cas9 mediated genome editing in farm animals].Yi Chuan. 2016 Mar;38(3):217-26. doi: 10.16288/j.yczz.15-398. Yi Chuan. 2016. PMID: 27001476 Review. Chinese.
Cited by
-
Model organisms: beyond the inner circle.Nat Methods. 2013 Jun;10(6):471-3. doi: 10.1038/nmeth.2484. Nat Methods. 2013. PMID: 23722204 No abstract available.
-
Exogenous enzymes upgrade transgenesis and genetic engineering of farm animals.Cell Mol Life Sci. 2015 May;72(10):1907-29. doi: 10.1007/s00018-015-1842-1. Epub 2015 Feb 1. Cell Mol Life Sci. 2015. PMID: 25636347 Free PMC article. Review.
-
CRISPR-Based Genome Editing as a New Therapeutic Tool in Retinal Diseases.Mol Biotechnol. 2021 Sep;63(9):768-779. doi: 10.1007/s12033-021-00345-4. Epub 2021 May 31. Mol Biotechnol. 2021. PMID: 34057656 Review.
-
Is gene editing an acceptable alternative to castration in pigs?PLoS One. 2019 Jun 24;14(6):e0218176. doi: 10.1371/journal.pone.0218176. eCollection 2019. PLoS One. 2019. PMID: 31233520 Free PMC article.
-
Live pigs produced from genome edited zygotes.Sci Rep. 2013 Oct 10;3:2847. doi: 10.1038/srep02847. Sci Rep. 2013. PMID: 24108318 Free PMC article.
References
-
- Arnould S, Delenda C, Grizot S, Desseaux C, Pâques F, Silva GH, et al. The I-CreI meganuclease and its engineered derivatives: applications from cell modification to gene therapy. Protein Engineering Design and Selection. 2011;24:27–31. - PubMed
-
- Baguisi A, Behboodi E, Melican DT, Pollock JS, Destrempes MM, Cammuso C, et al. Production of goats by somatic cell nuclear transfer. Nature Biotechnology. 1999;17:456–461. - PubMed
-
- Baldassarre H, Wang B, Kafidi N, Gauthier M, Neveu N, Lapointe J, et al. Production of transgenic goats by pronuclear microinjection of in vitro produced zygotes derived from oocytes recovered by laparoscopy. Theriogenology. 2003;59:831–839. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

