Epigenetic regulation of condensin-mediated genome organization during the cell cycle and upon DNA damage through histone H3 lysine 56 acetylation
- PMID: 23084836
- PMCID: PMC3513591
- DOI: 10.1016/j.molcel.2012.09.011
Epigenetic regulation of condensin-mediated genome organization during the cell cycle and upon DNA damage through histone H3 lysine 56 acetylation
Abstract
Complex genome organizations participate in various nuclear processes including transcription, DNA replication, and repair. However, the mechanisms that generate and regulate these functional genome structures remain largely unknown. Here, we describe how the Ku heterodimer complex, which functions in nonhomologous end joining, mediates clustering of long terminal repeat retrotransposons at centromeres in fission yeast. We demonstrate that the CENP-B subunit, Abp1, functions as a recruiter of the Ku complex, which in turn loads the genome-organizing machinery condensin to retrotransposons. Intriguingly, histone H3 lysine 56 (H3K56) acetylation, which functions in DNA replication and repair, interferes with Ku localization at retrotransposons without disrupting Abp1 localization and, as a consequence, dissociates condensin from retrotransposons. This dissociation releases condensin-mediated genomic associations during S phase and upon DNA damage. ATR (ATM- and Rad3-related) kinase mediates the DNA damage response of condensin-mediated genome organization. Our study describes a function of H3K56 acetylation that neutralizes condensin-mediated genome organization.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
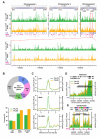
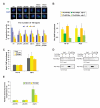
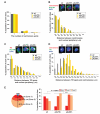
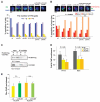

Comment in
-
A chromatin switch for chromosome condensation.Dev Cell. 2012 Dec 11;23(6):1127-8. doi: 10.1016/j.devcel.2012.11.016. Dev Cell. 2012. PMID: 23237949
Similar articles
-
Cohesin Causes Replicative DNA Damage by Trapping DNA Topological Stress.Mol Cell. 2020 May 21;78(4):739-751.e8. doi: 10.1016/j.molcel.2020.03.013. Epub 2020 Apr 6. Mol Cell. 2020. PMID: 32259483 Free PMC article.
-
Yeast histone H3 lysine 4 demethylase Jhd2 regulates mitotic rDNA condensation.BMC Biol. 2014 Sep 24;12:75. doi: 10.1186/s12915-014-0075-3. BMC Biol. 2014. PMID: 25248920 Free PMC article.
-
Histone H3 Thr 45 phosphorylation is a replication-associated post-translational modification in S. cerevisiae.Nat Cell Biol. 2010 Mar;12(3):294-8. doi: 10.1038/ncb2030. Epub 2010 Feb 7. Nat Cell Biol. 2010. PMID: 20139971 Free PMC article.
-
The Yeast Genomes in Three Dimensions: Mechanisms and Functions.Annu Rev Genet. 2017 Nov 27;51:23-44. doi: 10.1146/annurev-genet-120116-023438. Epub 2017 Aug 30. Annu Rev Genet. 2017. PMID: 28853923 Review.
-
Potential roles of condensin in genome organization and beyond in fission yeast.J Microbiol. 2021 May;59(5):449-459. doi: 10.1007/s12275-021-1039-2. Epub 2021 Apr 20. J Microbiol. 2021. PMID: 33877578 Review.
Cited by
-
Condensin action and compaction.Curr Genet. 2019 Apr;65(2):407-415. doi: 10.1007/s00294-018-0899-4. Epub 2018 Oct 25. Curr Genet. 2019. PMID: 30361853 Free PMC article. Review.
-
Involvement of condensin-directed gene associations in the organization and regulation of chromosome territories during the cell cycle.Nucleic Acids Res. 2016 May 5;44(8):3618-28. doi: 10.1093/nar/gkv1502. Epub 2015 Dec 23. Nucleic Acids Res. 2016. PMID: 26704981 Free PMC article.
-
Chromosome domain architecture and dynamic organization of the fission yeast genome.FEBS Lett. 2015 Oct 7;589(20 Pt A):2975-86. doi: 10.1016/j.febslet.2015.06.008. Epub 2015 Jun 19. FEBS Lett. 2015. PMID: 26096785 Free PMC article. Review.
-
Condensin-mediated chromosome organization in fission yeast.Curr Genet. 2016 Nov;62(4):739-743. doi: 10.1007/s00294-016-0601-7. Epub 2016 Apr 9. Curr Genet. 2016. PMID: 27061734 Free PMC article. Review.
-
Dose- and time-dependent epigenetic changes in the livers of Fisher 344 rats exposed to furan.Toxicol Sci. 2014 Jun;139(2):371-80. doi: 10.1093/toxsci/kfu044. Epub 2014 Mar 10. Toxicol Sci. 2014. PMID: 24614236 Free PMC article.
References
-
- Bähler J, Wu J, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. - PubMed
-
- Cam HP, Noma K, Ebina H, Levin HL, Grewal SI. Host genome surveillance for retrotransposons by transposon-derived proteins. Nature. 2008;451:431–436. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

