Isotope labeling for solution and solid-state NMR spectroscopy of membrane proteins
- PMID: 23076578
- PMCID: PMC3555569
- DOI: 10.1007/978-94-007-4954-2_3
Isotope labeling for solution and solid-state NMR spectroscopy of membrane proteins
Abstract
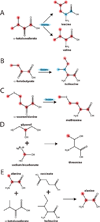
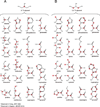
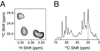
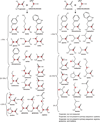
In this chapter, we summarize the isotopic labeling strategies used to obtain high-quality solution and solid-state NMR spectra of biological samples, with emphasis on integral membrane proteins (IMPs). While solution NMR is used to study IMPs under fast tumbling conditions, such as in the presence of detergent micelles or isotropic bicelles, solid-state NMR is used to study the structure and orientation of IMPs in lipid vesicles and bilayers. In spite of the tremendous progress in biomolecular NMR spectroscopy, the homogeneity and overall quality of the sample is still a substantial obstacle to overcome. Isotopic labeling is a major avenue to simplify overlapped spectra by either diluting the NMR active nuclei or allowing the resonances to be separated in multiple dimensions. In the following we will discuss isotopic labeling approaches that have been successfully used in the study of IMPs by solution and solid-state NMR spectroscopy.
Figures





Similar articles
-
From Nanodiscs to Isotropic Bicelles: A Procedure for Solution Nuclear Magnetic Resonance Studies of Detergent-Sensitive Integral Membrane Proteins.Structure. 2016 Oct 4;24(10):1830-1841. doi: 10.1016/j.str.2016.07.017. Epub 2016 Sep 15. Structure. 2016. PMID: 27618661 Free PMC article.
-
Efficient Segmental Isotope Labeling of Integral Membrane Proteins for High-Resolution NMR Studies.J Am Chem Soc. 2024 Jun 5;146(22):15403-15410. doi: 10.1021/jacs.4c03294. Epub 2024 May 24. J Am Chem Soc. 2024. PMID: 38787792 Free PMC article.
-
Isotope labeling of eukaryotic membrane proteins in yeast for solid-state NMR.Methods Enzymol. 2015;565:193-212. doi: 10.1016/bs.mie.2015.05.010. Epub 2015 Jun 18. Methods Enzymol. 2015. PMID: 26577733
-
Solution NMR spectroscopy of membrane proteins.Biochim Biophys Acta Biomembr. 2020 Sep 1;1862(9):183356. doi: 10.1016/j.bbamem.2020.183356. Epub 2020 May 13. Biochim Biophys Acta Biomembr. 2020. PMID: 32416193 Review.
-
Methyl-specific isotopic labeling: a molecular tool box for solution NMR studies of large proteins.Curr Opin Struct Biol. 2015 Jun;32:113-22. doi: 10.1016/j.sbi.2015.03.009. Epub 2015 Apr 13. Curr Opin Struct Biol. 2015. PMID: 25881211 Review.
Cited by
-
Nuclear spin effects in biological processes.Proc Natl Acad Sci U S A. 2023 Aug 8;120(32):e2300828120. doi: 10.1073/pnas.2300828120. Epub 2023 Jul 31. Proc Natl Acad Sci U S A. 2023. PMID: 37523549 Free PMC article.
-
Structural dynamics and topology of phosphorylated phospholamban homopentamer reveal its role in the regulation of calcium transport.Structure. 2013 Dec 3;21(12):2119-30. doi: 10.1016/j.str.2013.09.008. Epub 2013 Oct 24. Structure. 2013. PMID: 24207128 Free PMC article.
-
Improved spectral resolution of [13C,1H]-HSQC spectra of aromatic amino acid residues in proteins produced by cell-free synthesis from inexpensive 13C-labelled precursors.J Biomol NMR. 2023 Aug;77(4):183-190. doi: 10.1007/s10858-023-00420-9. Epub 2023 Jun 20. J Biomol NMR. 2023. PMID: 37338652 Free PMC article.
-
Solid state NMR of isotope labelled murine fur: a powerful tool to study atomic level keratin structure and treatment effects.J Biomol NMR. 2016 Oct;66(2):93-98. doi: 10.1007/s10858-016-0056-7. Epub 2016 Oct 3. J Biomol NMR. 2016. PMID: 27699524 Free PMC article.
-
Non-Canonical Amino Acids in Analyses of Protease Structure and Function.Int J Mol Sci. 2023 Sep 13;24(18):14035. doi: 10.3390/ijms241814035. Int J Mol Sci. 2023. PMID: 37762340 Free PMC article. Review.
References
-
- Crespi HL, Katz JJ. High resolution proton magnetic resonance studies of fully deuterated and isotope hybrid proteins. Nature. 1969;224:560–562. - PubMed
-
- Crespi HL, Rosenberg RM, Katz JJ. Proton magnetic resonance of proteins fully deuterated except for 1H-leucine side chains. Science. 1968;161:795–796. - PubMed
-
- Markley JL, Putter I, Jardetzky O. High-resolution nuclear magnetic resonance spectra of selectively deuterated staphylococcal nuclease. Science. 1968;161:1249–1251. - PubMed
-
- Ohki S, Kainosho M. Stable isotope labeling methods for protein NMR spectroscopy. Prog Nucl Magn Reson Spectrosc. 2008;53:208–226.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

