Signal amplification by glyco-qPCR for ultrasensitive detection of carbohydrates: applications in glycobiology
- PMID: 23073897
- PMCID: PMC3544480
- DOI: 10.1002/anie.201205112
Signal amplification by glyco-qPCR for ultrasensitive detection of carbohydrates: applications in glycobiology
Abstract
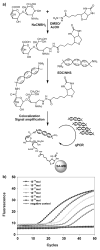
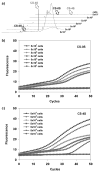
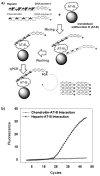
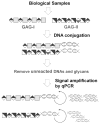
Tiny amounts of carbohydrates (ca. 1 zmol) can be detected quantitatively by a real-time method based on the conjugation of carbohydrates with DNA markers (see picture). The proposed method (glyco-qPCR) provides uniform, ultrasensitive detection of carbohydrates, which can be applied to glycobiology, as well as carbohydrate-based drug discovery.
Figures
Similar articles
-
A Glyco-mix of Approaches To Studying Carbohydrates.ACS Chem Biol. 2009 Sep 18;4(9):685-6. doi: 10.1021/cb900222x. ACS Chem Biol. 2009. PMID: 19761247 No abstract available.
-
Highlight: dolce vita--a sample from European glycobiology.Biol Chem. 2012 Aug;393(8):659. doi: 10.1515/hsz-2012-0232. Biol Chem. 2012. PMID: 22944670 No abstract available.
-
Glycobiology: A spoonful of sugar.Nature. 2009 Jan 29;457(7229):617-20. doi: 10.1038/457617a. Nature. 2009. PMID: 19177129 No abstract available.
-
Carbohydrate microarrays: key developments in glycobiology.Biol Chem. 2009 Jul;390(7):647-56. doi: 10.1515/BC.2009.071. Biol Chem. 2009. PMID: 19426131 Review.
-
40 years of glyco-polyacrylamide in glycobiology.Glycoconj J. 2021 Feb;38(1):89-100. doi: 10.1007/s10719-020-09965-5. Epub 2021 Jan 14. Glycoconj J. 2021. PMID: 33443721 Review.
Cited by
-
Recent advances in biotechnology for heparin and heparan sulfate analysis.Talanta. 2020 Nov 1;219:121270. doi: 10.1016/j.talanta.2020.121270. Epub 2020 Jun 14. Talanta. 2020. PMID: 32887160 Free PMC article. Review.
-
Applying DNA rolling circle amplification in fluorescence imaging of cell surface glycans labeled by a metabolic method.Chem Sci. 2016 Sep 1;7(9):6182-6189. doi: 10.1039/c6sc02089e. Epub 2016 Jun 14. Chem Sci. 2016. PMID: 30034758 Free PMC article.
-
Genetically encoded multivalent liquid glycan array displayed on M13 bacteriophage.Nat Chem Biol. 2021 Jul;17(7):806-816. doi: 10.1038/s41589-021-00788-5. Epub 2021 May 6. Nat Chem Biol. 2021. PMID: 33958792 Free PMC article.
-
High sensitivity detection of active botulinum neurotoxin by glyco-quantitative polymerase chain-reaction.Anal Chem. 2014 Mar 4;86(5):2279-84. doi: 10.1021/ac500262d. Epub 2014 Feb 24. Anal Chem. 2014. PMID: 24506443 Free PMC article.
-
Chemoenzymatic synthesis of glycosaminoglycans: re-creating, re-modeling and re-designing nature's longest or most complex carbohydrate chains.Glycobiology. 2013 Jul;23(7):764-77. doi: 10.1093/glycob/cwt016. Epub 2013 Mar 11. Glycobiology. 2013. PMID: 23481097 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

