ANALYSIS OF ALEXANDRIUM TAMARENSE (DINOPHYCEAE) GENES REVEALS THE COMPLEX EVOLUTIONARY HISTORY OF A MICROBIAL EUKARYOTE()
- PMID: 23066170
- PMCID: PMC3466611
- DOI: 10.1111/j.1529-8817.2012.01194.x
ANALYSIS OF ALEXANDRIUM TAMARENSE (DINOPHYCEAE) GENES REVEALS THE COMPLEX EVOLUTIONARY HISTORY OF A MICROBIAL EUKARYOTE()
Abstract
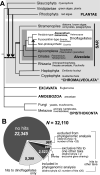
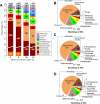
Microbial eukaryotes may extinguish much of their nuclear phylogenetic history due to endosymbiotic/horizontal gene transfer (E/HGT). We studied E/HGT in 32,110 contigs of expressed sequence tags (ESTs) from the dinoflagellate Alexandrium tamarense (Dinophyceae) using a conservative phylogenomic approach. The vast majority of predicted proteins (86.4%) in this alga are novel or dinoflagellate-specific. We searched for putative homologs of these predicted proteins against a taxonomically broadly sampled protein database that includes all currently available data from algae and protists and reconstructed a phylogeny from each of the putative homologous protein sets. Of the 2,523 resulting phylogenies, 14-17% are potentially impacted by E/HGT involving both prokaryote and eukaryote lineages, with 2-4% showing clear evidence of reticulate evolution. The complex evolutionary histories of the remaining proteins, many of which may also have been affected by E/HGT, cannot be interpreted using our approach with currently available gene data. We present empirical evidence of reticulate genome evolution that combined with inadequate or highly complex phylogenetic signal in many proteins may impede genome-wide approaches to infer the tree of microbial eukaryotes.
Figures


Similar articles
-
Horizontal gene transfer is a significant driver of gene innovation in dinoflagellates.Genome Biol Evol. 2013;5(12):2368-81. doi: 10.1093/gbe/evt179. Genome Biol Evol. 2013. PMID: 24259313 Free PMC article.
-
The impact of automated filtering of BLAST-determined homologs in the phylogenetic detection of horizontal gene transfer from a transcriptome assembly.Mol Phylogenet Evol. 2014 Feb;71:184-92. doi: 10.1016/j.ympev.2013.11.016. Epub 2013 Dec 7. Mol Phylogenet Evol. 2014. PMID: 24321593
-
Red and green algal origin of diatom membrane transporters: insights into environmental adaptation and cell evolution.PLoS One. 2011;6(12):e29138. doi: 10.1371/journal.pone.0029138. Epub 2011 Dec 14. PLoS One. 2011. PMID: 22195008 Free PMC article.
-
Horizontal gene transfer between microbial eukaryotes.Methods Mol Biol. 2009;532:473-87. doi: 10.1007/978-1-60327-853-9_27. Methods Mol Biol. 2009. PMID: 19271202 Review.
-
Horizontal Gene Transfer in Eukaryotes: Not if, but How Much?Trends Genet. 2020 Dec;36(12):915-925. doi: 10.1016/j.tig.2020.08.006. Epub 2020 Oct 1. Trends Genet. 2020. PMID: 33012528 Review.
Cited by
-
Core genes in diverse dinoflagellate lineages include a wealth of conserved dark genes with unknown functions.Sci Rep. 2018 Nov 21;8(1):17175. doi: 10.1038/s41598-018-35620-z. Sci Rep. 2018. PMID: 30464192 Free PMC article.
-
Single cell genome analysis of an uncultured heterotrophic stramenopile.Sci Rep. 2014 Apr 24;4:4780. doi: 10.1038/srep04780. Sci Rep. 2014. PMID: 24759094 Free PMC article.
-
Horizontal gene transfer is a significant driver of gene innovation in dinoflagellates.Genome Biol Evol. 2013;5(12):2368-81. doi: 10.1093/gbe/evt179. Genome Biol Evol. 2013. PMID: 24259313 Free PMC article.
-
Endosymbiotic gene transfer in tertiary plastid-containing dinoflagellates.Eukaryot Cell. 2014 Feb;13(2):246-55. doi: 10.1128/EC.00299-13. Epub 2013 Dec 2. Eukaryot Cell. 2014. PMID: 24297445 Free PMC article.
-
The eukaryotic tree of life from a global phylogenomic perspective.Cold Spring Harb Perspect Biol. 2014 May 1;6(5):a016147. doi: 10.1101/cshperspect.a016147. Cold Spring Harb Perspect Biol. 2014. PMID: 24789819 Free PMC article. Review.
References
-
- Baurain D, Brinkmann H, Petersen J, Rodríguez-Ezpeleta N, Stechmann A, Demoulin V, Roger AJ, Burger G, Lang BF, Philippe H. Phylogenomic evidence for separate acquisition of plastids in cryptophytes, haptophytes, and stramenopiles. Mol. Biol. Evol. 2010;27:1698–709. - PubMed
-
- Bergsten J. A review of long-branch attraction. Cladistics. 2005;21:163–93. - PubMed
-
- Bock R. The give-and-take of DNA: horizontal gene transfer in plants. Trends Plant Sci. 2010;15:11–22. - PubMed
-
- Bodył A, Mackiewicz P, Stiller JW. Early steps in plastid evolution: current ideas and controversies. BioEssays. 2009;31:1219–32. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

