UvrD Participation in Nucleotide Excision Repair Is Required for the Recovery of DNA Synthesis following UV-Induced Damage in Escherichia coli
- PMID: 23056919
- PMCID: PMC3465929
- DOI: 10.1155/2012/271453
UvrD Participation in Nucleotide Excision Repair Is Required for the Recovery of DNA Synthesis following UV-Induced Damage in Escherichia coli
Abstract
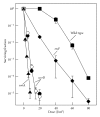
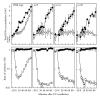
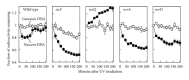
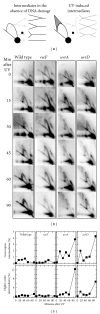
UvrD is a DNA helicase that participates in nucleotide excision repair and several replication-associated processes, including methyl-directed mismatch repair and recombination. UvrD is capable of displacing oligonucleotides from synthetic forked DNA structures in vitro and is essential for viability in the absence of Rep, a helicase associated with processing replication forks. These observations have led others to propose that UvrD may promote fork regression and facilitate resetting of the replication fork following arrest. However, the molecular activity of UvrD at replication forks in vivo has not been directly examined. In this study, we characterized the role UvrD has in processing and restoring replication forks following arrest by UV-induced DNA damage. We show that UvrD is required for DNA synthesis to recover. However, in the absence of UvrD, the displacement and partial degradation of the nascent DNA at the arrested fork occur normally. In addition, damage-induced replication intermediates persist and accumulate in uvrD mutants in a manner that is similar to that observed in other nucleotide excision repair mutants. These data indicate that, following arrest by DNA damage, UvrD is not required to catalyze fork regression in vivo and suggest that the failure of uvrD mutants to restore DNA synthesis following UV-induced arrest relates to its role in nucleotide excision repair.
Figures
Similar articles
-
Cellular characterization of the primosome and rep helicase in processing and restoration of replication following arrest by UV-induced DNA damage in Escherichia coli.J Bacteriol. 2012 Aug;194(15):3977-86. doi: 10.1128/JB.00290-12. Epub 2012 May 25. J Bacteriol. 2012. PMID: 22636770 Free PMC article.
-
lon incompatibility associated with mutations causing SOS induction: null uvrD alleles induce an SOS response in Escherichia coli.J Bacteriol. 2000 Jun;182(11):3151-7. doi: 10.1128/JB.182.11.3151-3157.2000. J Bacteriol. 2000. PMID: 10809694 Free PMC article.
-
Unwinding of forked DNA structures by UvrD.J Mol Biol. 2006 Sep 8;362(1):18-25. doi: 10.1016/j.jmb.2006.06.032. Epub 2006 Jun 30. J Mol Biol. 2006. PMID: 16890954
-
The Escherichia coli UvrD helicase is essential for Tus removal during recombination-dependent replication restart from Ter sites.Mol Microbiol. 2006 Oct;62(2):382-96. doi: 10.1111/j.1365-2958.2006.05382.x. Mol Microbiol. 2006. PMID: 17020578
-
Rescue of arrested replication forks by homologous recombination.Proc Natl Acad Sci U S A. 2001 Jul 17;98(15):8181-8. doi: 10.1073/pnas.111008798. Proc Natl Acad Sci U S A. 2001. PMID: 11459951 Free PMC article. Review.
Cited by
-
Molecular characterization of Paenibacillus antarcticus IPAC21, a bioemulsifier producer isolated from Antarctic soil.Front Microbiol. 2023 Mar 21;14:1142582. doi: 10.3389/fmicb.2023.1142582. eCollection 2023. Front Microbiol. 2023. PMID: 37025627 Free PMC article.
-
The CJIE1 prophage of Campylobacter jejuni affects protein expression in growth media with and without bile salts.BMC Microbiol. 2014 Mar 19;14:70. doi: 10.1186/1471-2180-14-70. BMC Microbiol. 2014. PMID: 24641125 Free PMC article.
-
Temporal Gene Expression of the Cyanobacterium Arthrospira in Response to Gamma Rays.PLoS One. 2015 Aug 26;10(8):e0135565. doi: 10.1371/journal.pone.0135565. eCollection 2015. PLoS One. 2015. PMID: 26308624 Free PMC article.
-
Fluoroquinolone Persistence in Escherichia coli Requires DNA Repair despite Differing between Starving Populations.Microorganisms. 2022 Jan 26;10(2):286. doi: 10.3390/microorganisms10020286. Microorganisms. 2022. PMID: 35208744 Free PMC article.
-
Limited Capacity or Involvement of Excision Repair, Double-Strand Breaks, or Translesion Synthesis for Psoralen Cross-Link Repair in Escherichia coli.Genetics. 2018 Sep;210(1):99-112. doi: 10.1534/genetics.118.301239. Epub 2018 Jul 25. Genetics. 2018. PMID: 30045856 Free PMC article.
References
-
- Chan GL, Doetsch PW, Haseltine WA. Cyclobutane pyrimidine dimers and (6–4) photoproducts Block polymerization by DNA polymerase I. Biochemistry. 1985;24(21):5723–5728. - PubMed
-
- Lippke JA, Gordon LK, Brash DE, Haseltine WA. Distribution of UV light-induced damage in a defined sequence of human DNA: detection of alkaline-sensitive lesions at pyrimidine nucleoside-cytidine sequences. Proceedings of the National Academy of Sciences of the United States of America. 1981;78(6):3388–3392. - PMC - PubMed
-
- Chow KH, Courcelle J. RecO acts with RecF and RecR to protect and maintain replication forks blocked by UV-induced DNA damage in Escherichia coli . Journal of Biological Chemistry. 2004;279(5):3492–3496. - PubMed
-
- Courcelle J, Hanawalt PC. RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated Escherichia coli . Molecular and General Genetics. 1999;262(3):543–551. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

