Nucleotide excision repair, mismatch repair, and R-loops modulate convergent transcription-induced cell death and repeat instability
- PMID: 23056461
- PMCID: PMC3463551
- DOI: 10.1371/journal.pone.0046807
Nucleotide excision repair, mismatch repair, and R-loops modulate convergent transcription-induced cell death and repeat instability
Abstract
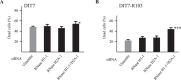
Expansion of CAG•CTG tracts located in specific genes is responsible for 13 human neurodegenerative disorders, the pathogenic mechanisms of which are not yet well defined. These disease genes are ubiquitously expressed in human tissues, and transcription has been identified as one of the major pathways destabilizing the repeats. Transcription-induced repeat instability depends on transcription-coupled nucleotide excision repair (TC-NER), the mismatch repair (MMR) recognition component MSH2/MSH3, and RNA/DNA hybrids (R-loops). Recently, we reported that simultaneous sense and antisense transcription-convergent transcription-through a CAG repeat not only promotes repeat instability, but also induces a cell stress response, which arrests the cell cycle and eventually leads to massive cell death via apoptosis. Here, we use siRNA knockdowns to investigate whether NER, MMR, and R-loops also modulate convergent-transcription-induced cell death and repeat instability. We find that siRNA-mediated depletion of TC-NER components increases convergent transcription-induced cell death, as does the simultaneous depletion of RNase H1 and RNase H2A. In contrast, depletion of MSH2 decreases cell death. These results identify TC-NER, MMR recognition, and R-loops as modulators of convergent transcription-induced cell death and shed light on the molecular mechanism involved. We also find that the TC-NER pathway, MSH2, and R-loops modulate convergent transcription-induced repeat instability. These observations link the mechanisms of convergent transcription-induced repeat instability and convergent transcription-induced cell death, suggesting that a common structure may trigger both outcomes.
Conflict of interest statement
Figures

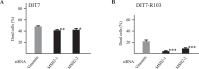
Similar articles
-
Mismatch repair enhances convergent transcription-induced cell death at trinucleotide repeats by activating ATR.DNA Repair (Amst). 2016 Jun;42:26-32. doi: 10.1016/j.dnarep.2016.03.016. Epub 2016 Apr 16. DNA Repair (Amst). 2016. PMID: 27131875 Free PMC article.
-
Diverse effects of individual mismatch repair components on transcription-induced CAG repeat instability in human cells.DNA Repair (Amst). 2009 Aug 6;8(8):878-85. doi: 10.1016/j.dnarep.2009.04.024. Epub 2009 Jun 3. DNA Repair (Amst). 2009. PMID: 19497791 Free PMC article.
-
Convergent transcription through a long CAG tract destabilizes repeats and induces apoptosis.Mol Cell Biol. 2010 Sep;30(18):4435-51. doi: 10.1128/MCB.00332-10. Epub 2010 Jul 20. Mol Cell Biol. 2010. PMID: 20647539 Free PMC article.
-
Disease-associated repeat instability and mismatch repair.DNA Repair (Amst). 2016 Feb;38:117-126. doi: 10.1016/j.dnarep.2015.11.008. Epub 2015 Dec 12. DNA Repair (Amst). 2016. PMID: 26774442 Review.
-
Transcription destabilizes triplet repeats.Mol Carcinog. 2009 Apr;48(4):350-61. doi: 10.1002/mc.20488. Mol Carcinog. 2009. PMID: 18973172 Free PMC article. Review.
Cited by
-
Twisting right to left: A…A mismatch in a CAG trinucleotide repeat overexpansion provokes left-handed Z-DNA conformation.PLoS Comput Biol. 2015 Apr 13;11(4):e1004162. doi: 10.1371/journal.pcbi.1004162. eCollection 2015 Apr. PLoS Comput Biol. 2015. PMID: 25876062 Free PMC article.
-
Repeat instability during DNA repair: Insights from model systems.Crit Rev Biochem Mol Biol. 2015 Mar-Apr;50(2):142-67. doi: 10.3109/10409238.2014.999192. Epub 2015 Jan 22. Crit Rev Biochem Mol Biol. 2015. PMID: 25608779 Free PMC article. Review.
-
R-loops associated with triplet repeat expansions promote gene silencing in Friedreich ataxia and fragile X syndrome.PLoS Genet. 2014 May 1;10(5):e1004318. doi: 10.1371/journal.pgen.1004318. eCollection 2014 May. PLoS Genet. 2014. PMID: 24787137 Free PMC article.
-
Mismatch repair enhances convergent transcription-induced cell death at trinucleotide repeats by activating ATR.DNA Repair (Amst). 2016 Jun;42:26-32. doi: 10.1016/j.dnarep.2016.03.016. Epub 2016 Apr 16. DNA Repair (Amst). 2016. PMID: 27131875 Free PMC article.
-
Conserved Senescence Associated Genes and Pathways in Primary Human Fibroblasts Detected by RNA-Seq.PLoS One. 2016 May 3;11(5):e0154531. doi: 10.1371/journal.pone.0154531. eCollection 2016. PLoS One. 2016. PMID: 27140416 Free PMC article.
References
-
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, et al. (2001) The sequence of the human genome. Science 291: 1304–1351. - PubMed
-
- Orr HT, Zoghbi HY (2007) Trinucleotide repeat disorders. Annu Rev Neurosci 30: 575–621. - PubMed
-
- Gatchel JR, Zoghbi HY (2005) Diseases of unstable repeat expansion: mechanisms and common principles. Nat Rev Genet 6: 743–755. - PubMed
-
- Pearson CE, Edamura KN, Cleary JD (2005) Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet 6: 729–742. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

