Signaling-dependent phosphorylation of mitotic centromere-associated kinesin regulates microtubule depolymerization and its centrosomal localization
- PMID: 23055517
- PMCID: PMC3504770
- DOI: 10.1074/jbc.M112.399576
Signaling-dependent phosphorylation of mitotic centromere-associated kinesin regulates microtubule depolymerization and its centrosomal localization
Abstract
Background: Although PAK1 regulates cytoskeleton and microtubule dynamics, its role in controlling the functions of MCAK remains unknown.
Results: PAK1 phosphorylates MCAK and thereby regulates both its localization and function.
Conclusion: MCAK is a cognate substrate of PAK1.
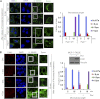
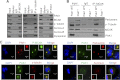
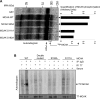
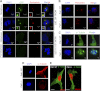
Significance: This study provides a novel mechanistic insight into PAK1 regulation of MCAK functions. Although p21-activated kinase 1 (PAK1) and microtubule (MT) dynamics regulate numerous fundamental processes including cytoskeleton remodeling, directional motility, and mitotic functions, the significance of PAK1 signaling in regulating the functions of MT-destabilizing protein mitotic centromere-associated kinesin (MCAK) remains unknown. Here we found that MCAK is a cognate substrate of PAK1 wherein PAK1 phosphorylates MCAK on serines 192 and 111 both in vivo and in vitro. Furthermore, we found that PAK1 phosphorylation of MCAK on serines 192 and 111 preferentially regulates its microtubule depolymerization activity and localization to centrosomes, respectively, in the mammalian cells.
Figures


Similar articles
-
The activity regulation of the mitotic centromere-associated kinesin by Polo-like kinase 1.Oncotarget. 2015 Mar 30;6(9):6641-55. doi: 10.18632/oncotarget.2843. Oncotarget. 2015. PMID: 25504441 Free PMC article.
-
Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity.Curr Biol. 2004 Feb 17;14(4):273-86. doi: 10.1016/j.cub.2004.01.055. Curr Biol. 2004. PMID: 14972678
-
Functional and spatial regulation of mitotic centromere-associated kinesin by cyclin-dependent kinase 1.Mol Cell Biol. 2010 Jun;30(11):2594-607. doi: 10.1128/MCB.00098-10. Epub 2010 Apr 5. Mol Cell Biol. 2010. PMID: 20368358 Free PMC article.
-
Mitotic centromere-associated kinesin (MCAK): a potential cancer drug target.Oncotarget. 2011 Dec;2(12):935-47. doi: 10.18632/oncotarget.416. Oncotarget. 2011. PMID: 22249213 Free PMC article. Review.
-
Molecular insight into the regulation and function of MCAK.Crit Rev Biochem Mol Biol. 2015 Jul-Aug;51(4):228-45. doi: 10.1080/10409238.2016.1178705. Epub 2016 May 5. Crit Rev Biochem Mol Biol. 2015. PMID: 27146484 Review.
Cited by
-
The KLP-7 Residue S546 Is a Putative Aurora Kinase Site Required for Microtubule Regulation at the Centrosome in C. elegans.PLoS One. 2015 Jul 13;10(7):e0132593. doi: 10.1371/journal.pone.0132593. eCollection 2015. PLoS One. 2015. PMID: 26168236 Free PMC article.
-
The activity regulation of the mitotic centromere-associated kinesin by Polo-like kinase 1.Oncotarget. 2015 Mar 30;6(9):6641-55. doi: 10.18632/oncotarget.2843. Oncotarget. 2015. PMID: 25504441 Free PMC article.
-
Structure, biochemistry, and biology of PAK kinases.Gene. 2017 Mar 20;605:20-31. doi: 10.1016/j.gene.2016.12.014. Epub 2016 Dec 19. Gene. 2017. PMID: 28007610 Free PMC article. Review.
-
Functional analysis of phosphorylation of the mitotic centromere-associated kinesin by Aurora B kinase in human tumor cells.Cell Cycle. 2015;14(23):3755-67. doi: 10.1080/15384101.2015.1068481. Epub 2015 Jul 6. Cell Cycle. 2015. PMID: 26148251 Free PMC article.
-
Spatiotemporal dynamics of Aurora B-PLK1-MCAK signaling axis orchestrates kinetochore bi-orientation and faithful chromosome segregation.Sci Rep. 2015 Jul 24;5:12204. doi: 10.1038/srep12204. Sci Rep. 2015. PMID: 26206521 Free PMC article.
References
-
- Maney T., Wagenbach M., Wordeman L. (2001) Molecular dissection of the microtubule depolymerizing activity of mitotic centromere-associated kinesin. J. Biol. Chem. 276, 34753–34758 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

