Structural insights into activation of antiviral NK cell responses
- PMID: 23046134
- PMCID: PMC3471384
- DOI: 10.1111/j.1600-065X.2012.01168.x
Structural insights into activation of antiviral NK cell responses
Abstract
Natural killer (NK) cells are key components of innate immune responses, providing surveillance against cells undergoing tumorigenesis or infection, by viruses or internal pathogens. NK cells can directly eliminate compromised cells and regulate downstream responses of the innate and acquired immune systems through the release of immune modulators (cytokines, interferons). The importance of the role NK cells play in immune defense was demonstrated originally in herpes viral infections, usually mild or localized, which become severe and life threatening in NK-deficient patients . NK cell effector functions are governed by balancing opposing signals from a diverse array of activating and inhibitory receptors. Many NK receptors occur in paired activating and inhibitory isoforms and recognize major histocompatibility complex (MHC) class I proteins with varying degrees of peptide specificity. Structural studies have made considerable inroads into understanding the molecular mechanisms employed to broadly recognize multiple MHC ligands or specific pathogen-associated antigens and the strategies employed by viruses to thwart these defenses. Although many details of NK development, signaling, and integration remain mysterious, it is clear that NK receptors are key components of a system exquisitely tuned to sense any dysregulation in MHC class I expression, or the expression of certain viral antigens, resulting in the elimination of affected cells.
© 2012 John Wiley & Sons A/S.
Figures


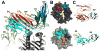
Similar articles
-
Natural killer cell responses during viral infections: flexibility and conditioning of innate immunity by experience.Curr Opin Virol. 2011 Dec;1(6):497-512. doi: 10.1016/j.coviro.2011.10.017. Curr Opin Virol. 2011. PMID: 22180766 Free PMC article. Review.
-
Viral evasion of natural killer cells.Nat Immunol. 2002 Nov;3(11):1006-12. doi: 10.1038/ni1102-1006. Nat Immunol. 2002. PMID: 12407408 Review.
-
Zika Virus Escapes NK Cell Detection by Upregulating Major Histocompatibility Complex Class I Molecules.J Virol. 2017 Oct 27;91(22):e00785-17. doi: 10.1128/JVI.00785-17. Print 2017 Nov 15. J Virol. 2017. PMID: 28878071 Free PMC article.
-
Viral Evasion of Natural Killer Cell Activation.Viruses. 2016 Apr 12;8(4):95. doi: 10.3390/v8040095. Viruses. 2016. PMID: 27077876 Free PMC article. Review.
-
Regulatory NK cells in autoimmune disease.J Autoimmun. 2012 Sep;39(3):206-15. doi: 10.1016/j.jaut.2012.05.006. Epub 2012 Jun 15. J Autoimmun. 2012. PMID: 22704425 Review.
Cited by
-
The structure of the atypical killer cell immunoglobulin-like receptor, KIR2DL4.J Biol Chem. 2015 Apr 17;290(16):10460-71. doi: 10.1074/jbc.M114.612291. Epub 2015 Mar 10. J Biol Chem. 2015. PMID: 25759384 Free PMC article.
-
Effects of HLA single chain trimer design on peptide presentation and stability.Front Immunol. 2023 May 3;14:1170462. doi: 10.3389/fimmu.2023.1170462. eCollection 2023. Front Immunol. 2023. PMID: 37207206 Free PMC article.
-
HLA-F and MHC-I Open Conformers Bind Natural Killer Cell Ig-Like Receptor KIR3DS1.PLoS One. 2016 Sep 20;11(9):e0163297. doi: 10.1371/journal.pone.0163297. eCollection 2016. PLoS One. 2016. PMID: 27649529 Free PMC article.
-
NK Cells during Dengue Disease and Their Recognition of Dengue Virus-Infected cells.Front Immunol. 2014 May 5;5:192. doi: 10.3389/fimmu.2014.00192. eCollection 2014. Front Immunol. 2014. PMID: 24829565 Free PMC article. Review.
-
Leukocyte Ig-Like Receptors - A Model for MHC Class I Disease Associations.Front Immunol. 2016 Jul 25;7:281. doi: 10.3389/fimmu.2016.00281. eCollection 2016. Front Immunol. 2016. PMID: 27504110 Free PMC article. Review.
References
-
- Biron CA, Byron KS, Sullivan JL. Severe herpesvirus infections in an adolescent without natural killer cells. N Engl J Med. 1989;320:1731–1735. - PubMed
-
- Bancroft GJ. The role of natural killer cells in innate resistance to infection. Curr Opin Immunol. 1993;5:503–510. - PubMed
-
- Biron CA, Nguyen KB, Pien GC, Cousens LP, Salazar-Mather TP. Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu Rev Immunol. 1999;17:189–220. - PubMed
-
- Cooper MA, Fehniger TA, Caligiuri MA. The biology of human natural killer-cell subsets. Trends Immunol. 2001;22:633–640. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

