Advancement of the 10-species subgingival Zurich biofilm model by examining different nutritional conditions and defining the structure of the in vitro biofilms
- PMID: 23040057
- PMCID: PMC3561252
- DOI: 10.1186/1471-2180-12-227
Advancement of the 10-species subgingival Zurich biofilm model by examining different nutritional conditions and defining the structure of the in vitro biofilms
Abstract
Background: Periodontitis is caused by a highly complex consortium of bacteria that establishes as biofilms in subgingival pockets. It is a disease that occurs worldwide and its consequences are a major health concern. Investigations in situ are not possible and the bacterial community varies greatly between patients and even within different loci. Due to the high complexity of the consortium and the availability of samples, a clear definition of the pathogenic bacteria and their mechanisms of pathogenicity are still not available. In the current study we addressed the need of a defined model system by advancing our previously described subgingival biofilm model towards a bacterial composition that reflects the one observed in diseased sites of patients and analysed the structure of these biofilms.
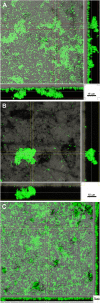
Results: We further developed the growth media by systematic variation of key components resulting in improved stability and the firm establishment of spirochetes in the 10-species subgingival Zurich biofilm model. A high concentration of heat-inactivated human serum allowed the best proliferation of the used species. Therefore we further investigated these biofilms by analysing their structure by confocal laser scanning microscopy following fluorescence in situ hybridisation. The species showed mutual interactions as expected from other studies. The abundances of all organisms present in this model were determined by microscopic counting following species-specific identification by both fluorescence in situ hybridisation and immunofluorescence. The newly integrated treponemes were the most abundant organisms.
Conclusions: The use of 50% of heat-inactivated human serum used in the improved growth medium resulted in significantly thicker and more stable biofilms, and the quantitative representation of the used species represents the in vivo community of periodontitis patients much closer than in biofilms grown in the two media with less or no human serum. The appearance of T. denticola, P. gingivalis, and T. forsythia in the top layer of the biofilms, and the high abundance of T. denticola, reflects well the microbial situation observed at diseased sites. The improved model biofilms will allow further investigations of interactions between individual species and of the effects of atmospheric or nutritional changes, as well as interactions with tissue cells.
Figures


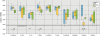


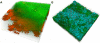


Similar articles
-
Validation of a quantitative real-time PCR assay and comparison with fluorescence microscopy and selective agar plate counting for species-specific quantification of an in vitro subgingival biofilm model.J Periodontal Res. 2013 Aug;48(4):517-26. doi: 10.1111/jre.12034. Epub 2012 Dec 30. J Periodontal Res. 2013. PMID: 23278531
-
Confocal Raman microscopy to identify bacteria in oral subgingival biofilm models.PLoS One. 2020 May 11;15(5):e0232912. doi: 10.1371/journal.pone.0232912. eCollection 2020. PLoS One. 2020. PMID: 32392236 Free PMC article.
-
Consistent and reproducible long-term in vitro growth of health and disease-associated oral subgingival biofilms.BMC Microbiol. 2018 Jul 11;18(1):70. doi: 10.1186/s12866-018-1212-x. BMC Microbiol. 2018. PMID: 29996764 Free PMC article.
-
Subgingival biofilm structure.Front Oral Biol. 2012;15:1-16. doi: 10.1159/000329667. Epub 2011 Nov 11. Front Oral Biol. 2012. PMID: 22142954 Review.
-
Microbial communities and their interactions in biofilm systems: an overview.Water Sci Technol. 2004;49(11-12):327-36. Water Sci Technol. 2004. PMID: 15303758 Review.
Cited by
-
Spatiotemporal monitoring of a periodontal multispecies biofilm model: demonstration of prebiotic treatment responses.Appl Environ Microbiol. 2023 Oct 31;89(10):e0108123. doi: 10.1128/aem.01081-23. Epub 2023 Sep 28. Appl Environ Microbiol. 2023. PMID: 37768099 Free PMC article.
-
Multispecies biofilm behavior and host interaction support the association of Tannerella serpentiformis with periodontal health.Mol Oral Microbiol. 2023 Apr;38(2):115-133. doi: 10.1111/omi.12385. Epub 2022 Aug 29. Mol Oral Microbiol. 2023. PMID: 35964247 Free PMC article.
-
Polymicrobial Interactions of Oral Microbiota: a Historical Review and Current Perspective.mBio. 2022 Jun 28;13(3):e0023522. doi: 10.1128/mbio.00235-22. Epub 2022 May 2. mBio. 2022. PMID: 35491817 Free PMC article. Review.
-
Mimicking biofilm formation and development: Recent progress in in vitro and in vivo biofilm models.iScience. 2021 Apr 17;24(5):102443. doi: 10.1016/j.isci.2021.102443. eCollection 2021 May 21. iScience. 2021. PMID: 34013169 Free PMC article. Review.
-
Modified SHI medium supports growth of a disease-state subgingival polymicrobial community in vitro.Mol Oral Microbiol. 2021 Feb;36(1):37-49. doi: 10.1111/omi.12323. Epub 2020 Dec 3. Mol Oral Microbiol. 2021. PMID: 33174294 Free PMC article.
References
-
- Marsh PD, Percival RS. The oral microflora - friend or foe? Can we decide? Int Dent J. 2006;56:233–239. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

