SPOC1 modulates DNA repair by regulating key determinants of chromatin compaction and DNA damage response
- PMID: 23034801
- PMCID: PMC3526275
- DOI: 10.1093/nar/gks868
SPOC1 modulates DNA repair by regulating key determinants of chromatin compaction and DNA damage response
Erratum in
-
SPOC1 modulates DNA repair by regulating key determinants of chromatin compaction and DNA damage response.Nucleic Acids Res. 2020 Sep 25;48(17):10013-10014. doi: 10.1093/nar/gkaa754. Nucleic Acids Res. 2020. PMID: 32890405 Free PMC article. No abstract available.
Abstract
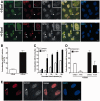
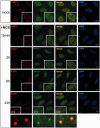
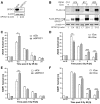
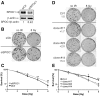
Survival time-associated plant homeodomain (PHD) finger protein in Ovarian Cancer 1 (SPOC1, also known as PHF13) is known to modulate chromatin structure and is essential for testicular stem-cell differentiation. Here we show that SPOC1 is recruited to DNA double-strand breaks (DSBs) in an ATM-dependent manner. Moreover, SPOC1 localizes at endogenous repair foci, including OPT domains and accumulates at large DSB repair foci characteristic for delayed repair at heterochromatic sites. SPOC1 depletion enhances the kinetics of ionizing radiation-induced foci (IRIF) formation after γ-irradiation (γ-IR), non-homologous end-joining (NHEJ) repair activity, and cellular radioresistance, but impairs homologous recombination (HR) repair. Conversely, SPOC1 overexpression delays IRIF formation and γH2AX expansion, reduces NHEJ repair activity and enhances cellular radiosensitivity. SPOC1 mediates dose-dependent changes in chromatin association of DNA compaction factors KAP-1, HP1-α and H3K9 methyltransferases (KMT) GLP, G9A and SETDB1. In addition, SPOC1 interacts with KAP-1 and H3K9 KMTs, inhibits KAP-1 phosphorylation and enhances H3K9 trimethylation. These findings provide the first evidence for a function of SPOC1 in DNA damage response (DDR) and repair. SPOC1 acts as a modulator of repair kinetics and choice of pathways. This involves its dose-dependent effects on DNA damage sensors, repair mediators and key regulators of chromatin structure.
Figures



Similar articles
-
The influence of heterochromatin on DNA double strand break repair: Getting the strong, silent type to relax.DNA Repair (Amst). 2010 Dec 10;9(12):1273-82. doi: 10.1016/j.dnarep.2010.09.013. Epub 2010 Oct 30. DNA Repair (Amst). 2010. PMID: 21036673 Review.
-
The impact of heterochromatin on DSB repair.Biochem Soc Trans. 2009 Jun;37(Pt 3):569-76. doi: 10.1042/BST0370569. Biochem Soc Trans. 2009. PMID: 19442252
-
53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair.Nat Cell Biol. 2010 Feb;12(2):177-84. doi: 10.1038/ncb2017. Epub 2010 Jan 17. Nat Cell Biol. 2010. PMID: 20081839
-
DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin.Proc Natl Acad Sci U S A. 2014 Jun 24;111(25):9169-74. doi: 10.1073/pnas.1403565111. Epub 2014 Jun 9. Proc Natl Acad Sci U S A. 2014. PMID: 24927542 Free PMC article.
-
Roles for 53BP1 in the repair of radiation-induced DNA double strand breaks.DNA Repair (Amst). 2020 Sep;93:102915. doi: 10.1016/j.dnarep.2020.102915. DNA Repair (Amst). 2020. PMID: 33087281 Review.
Cited by
-
KAP1 Is a Host Restriction Factor That Promotes Human Adenovirus E1B-55K SUMO Modification.J Virol. 2015 Nov 4;90(2):930-46. doi: 10.1128/JVI.01836-15. Print 2016 Jan 15. J Virol. 2015. PMID: 26537675 Free PMC article.
-
Discovery of a Novel Antiviral Effect of the Restriction Factor SPOC1 against Human Cytomegalovirus.Viruses. 2024 Feb 27;16(3):363. doi: 10.3390/v16030363. Viruses. 2024. PMID: 38543731 Free PMC article.
-
Loss of CHD1 causes DNA repair defects and enhances prostate cancer therapeutic responsiveness.EMBO Rep. 2016 Nov;17(11):1609-1623. doi: 10.15252/embr.201642352. Epub 2016 Sep 5. EMBO Rep. 2016. PMID: 27596623 Free PMC article.
-
Chromatin-Remodeling Factor SPOC1 Acts as a Cellular Restriction Factor against Human Cytomegalovirus by Repressing the Major Immediate Early Promoter.J Virol. 2018 Jun 29;92(14):e00342-18. doi: 10.1128/JVI.00342-18. Print 2018 Jul 15. J Virol. 2018. PMID: 29743358 Free PMC article.
-
Opposing ISWI- and CHD-class chromatin remodeling activities orchestrate heterochromatic DNA repair.J Cell Biol. 2014 Dec 22;207(6):717-33. doi: 10.1083/jcb.201405077. J Cell Biol. 2014. PMID: 25533843 Free PMC article.
References
-
- Mohrmann G, Hengstler JG, Hofmann TG, Endele SU, Lee B, Stelzer C, Zabel B, Brieger J, Hasenclever D, Tanner B, et al. SPOC1, a novel PHD-finger protein: association with residual disease and survival in ovarian cancer. Int. J. Cancer. 2005;116:547–554. - PubMed
-
- Kinkley S, Staege H, Mohrmann G, Rohaly G, Schaub T, Kremmer E, Winterpacht A, Will H. SPOC1: a novel PHD-containing protein modulating chromatin structure and mitotic chromosome condensation. J. Cell Sci. 2009;122:2946–2956. - PubMed
-
- Bordlein A, Scherthan H, Nelkenbrecher C, Molter T, Bosl MR, Dippold C, Birke K, Kinkley S, Staege H, Will H, et al. SPOC1 (PHF13) is required for spermatogonial stem cell differentiation and sustained spermatogenesis. J. Cell Sci. 2011;124:3137–3148. - PubMed
-
- Schuster-Bockler B, Lehner B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 2012;488:504–507. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

