Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models
- PMID: 23033316
- PMCID: PMC3558800
- DOI: 10.1002/humu.22225
Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models
Abstract
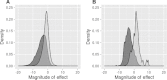
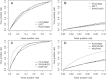
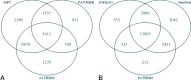
The rate at which nonsynonymous single nucleotide polymorphisms (nsSNPs) are being identified in the human genome is increasing dramatically owing to advances in whole-genome/whole-exome sequencing technologies. Automated methods capable of accurately and reliably distinguishing between pathogenic and functionally neutral nsSNPs are therefore assuming ever-increasing importance. Here, we describe the Functional Analysis Through Hidden Markov Models (FATHMM) software and server: a species-independent method with optional species-specific weightings for the prediction of the functional effects of protein missense variants. Using a model weighted for human mutations, we obtained performance accuracies that outperformed traditional prediction methods (i.e., SIFT, PolyPhen, and PANTHER) on two separate benchmarks. Furthermore, in one benchmark, we achieve performance accuracies that outperform current state-of-the-art prediction methods (i.e., SNPs&GO and MutPred). We demonstrate that FATHMM can be efficiently applied to high-throughput/large-scale human and nonhuman genome sequencing projects with the added benefit of phenotypic outcome associations. To illustrate this, we evaluated nsSNPs in wheat (Triticum spp.) to identify some of the important genetic variants responsible for the phenotypic differences introduced by intense selection during domestication. A Web-based implementation of FATHMM, including a high-throughput batch facility and a downloadable standalone package, is available at http://fathmm.biocompute.org.uk.
© 2012 Wiley Periodicals, Inc.
Figures
Similar articles
-
Ranking non-synonymous single nucleotide polymorphisms based on disease concepts.Hum Genomics. 2014 Jun 30;8(1):11. doi: 10.1186/1479-7364-8-11. Hum Genomics. 2014. PMID: 24980617 Free PMC article.
-
Predicting the functional consequences of cancer-associated amino acid substitutions.Bioinformatics. 2013 Jun 15;29(12):1504-10. doi: 10.1093/bioinformatics/btt182. Epub 2013 Apr 25. Bioinformatics. 2013. PMID: 23620363 Free PMC article.
-
A bioinformatics approach for the phenotype prediction of nonsynonymous single nucleotide polymorphisms in human cytochromes P450.Drug Metab Dispos. 2009 May;37(5):977-91. doi: 10.1124/dmd.108.026047. Epub 2009 Feb 9. Drug Metab Dispos. 2009. PMID: 19204079
-
Computational prediction of the effects of non-synonymous single nucleotide polymorphisms in human DNA repair genes.Neuroscience. 2007 Apr 14;145(4):1273-9. doi: 10.1016/j.neuroscience.2006.09.004. Epub 2006 Oct 19. Neuroscience. 2007. PMID: 17055652 Review.
-
Predicting the effects of amino acid substitutions on protein function.Annu Rev Genomics Hum Genet. 2006;7:61-80. doi: 10.1146/annurev.genom.7.080505.115630. Annu Rev Genomics Hum Genet. 2006. PMID: 16824020 Review.
Cited by
-
m6A-TSHub: Unveiling the Context-specific m6A Methylation and m6A-affecting Mutations in 23 Human Tissues.Genomics Proteomics Bioinformatics. 2023 Aug;21(4):678-694. doi: 10.1016/j.gpb.2022.09.001. Epub 2022 Sep 9. Genomics Proteomics Bioinformatics. 2023. PMID: 36096444 Free PMC article.
-
Whole-Exome Screening and Analysis of Signaling Pathways in Multiple Endocrine Neoplasia Type 1 Patients with Different Outcomes: Insights into Cellular Mechanisms and Possible Functional Implications.Int J Mol Sci. 2024 Jan 15;25(2):1065. doi: 10.3390/ijms25021065. Int J Mol Sci. 2024. PMID: 38256138 Free PMC article.
-
Genes with de novo mutations are shared by four neuropsychiatric disorders discovered from NPdenovo database.Mol Psychiatry. 2016 Feb;21(2):290-7. doi: 10.1038/mp.2015.40. Epub 2015 Apr 7. Mol Psychiatry. 2016. PMID: 25849321 Free PMC article.
-
A novel SNCA E83Q mutation in a case of dementia with Lewy bodies and atypical frontotemporal lobar degeneration.Neuropathology. 2020 Dec;40(6):620-626. doi: 10.1111/neup.12687. Epub 2020 Aug 12. Neuropathology. 2020. PMID: 32786148 Free PMC article.
-
A pan-cancer analysis reveals nonstop extension mutations causing SMAD4 tumour suppressor degradation.Nat Cell Biol. 2020 Aug;22(8):999-1010. doi: 10.1038/s41556-020-0551-7. Epub 2020 Jul 27. Nat Cell Biol. 2020. PMID: 32719554
References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

