Infectivity-selective oncolytic adenovirus developed by high-throughput screening of adenovirus-formatted library
- PMID: 23032977
- PMCID: PMC3538312
- DOI: 10.1038/mt.2012.205
Infectivity-selective oncolytic adenovirus developed by high-throughput screening of adenovirus-formatted library
Abstract
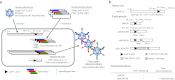
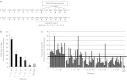
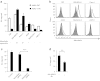
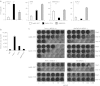
Adenovirus (Ad) is a potent gene-delivery vehicle and has frequently been used for designing oncolytic viruses. However, lack of selectivity on infection has hampered the achievement of sufficient in vivo efficiency. Here, we developed a novel oncolytic virus system, infectivity-selective oncolytic adenovirus (ISOAd), via direct high-throughput screening of a high-diversity targeting-ligand library in adenoviral format. Through our newly designed rescue virus system, the high-diversity Ad library carrying the random seven amino acid sequences ligand-library in the AB-loop of its fiber-knob region (5 × 10(9) diversity) was successfully generated. During the screening of this library with the cells expressing the target molecule (mesothelin, MSLN), the AB-loop sequence of the virus clones converged to one dominant sequence and a novel MSLN-targeting sequence was isolated. The virus with the isolated motif showed selective infectivity to MSLN-positive cells in vitro. In vivo, it exhibited a selective and potent antitumor effect resulted from the viral replication in MSLN-positive xenografts. The ISOAd is a novel class of oncolytic Ad, which has selectivity at the step of transduction. The selectivity at the stage of infection can open new perspectives in oncolytic Ad therapy for various diseases.
Figures



Similar articles
-
Oncolytic virus therapy for pancreatic cancer using the adenovirus library displaying random peptides on the fiber knob.Gene Ther. 2009 May;16(5):669-80. doi: 10.1038/gt.2009.1. Epub 2009 Feb 19. Gene Ther. 2009. PMID: 19225547
-
E1A, E1B double-restricted adenovirus with RGD-fiber modification exhibits enhanced oncolysis for CAR-deficient biliary cancers.Clin Cancer Res. 2007 May 15;13(10):3043-50. doi: 10.1158/1078-0432.CCR-06-2103. Clin Cancer Res. 2007. PMID: 17505007
-
A vesicular stomatitis virus glycoprotein epitope-incorporated oncolytic adenovirus overcomes CAR-dependency and shows markedly enhanced cancer cell killing and suppression of tumor growth.Oncotarget. 2015 Oct 27;6(33):34875-91. doi: 10.18632/oncotarget.5332. Oncotarget. 2015. PMID: 26430798 Free PMC article.
-
Tropism and transduction of oncolytic adenovirus 5 vectors in cancer therapy: Focus on fiber chimerism and mosaicism, hexon and pIX.Virus Res. 2018 Sep 15;257:40-51. doi: 10.1016/j.virusres.2018.08.012. Epub 2018 Aug 17. Virus Res. 2018. PMID: 30125593 Review.
-
The Development of Oncolytic Adenovirus Therapy in the Past and Future - For the Case of Pancreatic Cancer.Curr Cancer Drug Targets. 2018;18(2):153-161. doi: 10.2174/1568009617666170222123925. Curr Cancer Drug Targets. 2018. PMID: 28228084 Free PMC article. Review.
Cited by
-
Oncolytic viruses: From bench to bedside with a focus on safety.Hum Vaccin Immunother. 2015;11(7):1573-84. doi: 10.1080/21645515.2015.1037058. Hum Vaccin Immunother. 2015. PMID: 25996182 Free PMC article. Review.
-
Role of Gene Therapy in Pancreatic Cancer-A Review.Cancers (Basel). 2018 Apr 3;10(4):103. doi: 10.3390/cancers10040103. Cancers (Basel). 2018. PMID: 29614005 Free PMC article. Review.
-
Adenoviruses using the cancer marker EphA2 as a receptor in vitro and in vivo by genetic ligand insertion into different capsid scaffolds.PLoS One. 2014 Apr 23;9(4):e95723. doi: 10.1371/journal.pone.0095723. eCollection 2014. PLoS One. 2014. PMID: 24760010 Free PMC article.
-
Adenovirus infections in immunocompetent and immunocompromised patients.Clin Microbiol Rev. 2014 Jul;27(3):441-62. doi: 10.1128/CMR.00116-13. Clin Microbiol Rev. 2014. PMID: 24982316 Free PMC article. Review.
-
Intestinal Microbiota-A Promising Target for Antiviral Therapy?Front Immunol. 2021 May 12;12:676232. doi: 10.3389/fimmu.2021.676232. eCollection 2021. Front Immunol. 2021. PMID: 34054866 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

