Convergent transcription induces transcriptional gene silencing in fission yeast and mammalian cells
- PMID: 23022730
- PMCID: PMC3504457
- DOI: 10.1038/nsmb.2392
Convergent transcription induces transcriptional gene silencing in fission yeast and mammalian cells
Abstract
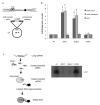
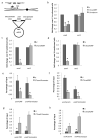
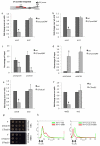
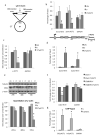
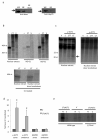
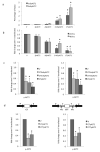
We show that convergent transcription induces transcriptional gene silencing (TGS) in trans for both fission yeast and mammalian cells. This method has advantages over existing strategies to induce gene silencing. Previous studies in fission yeast have characterized TGS as a cis-specific process involving RNA interference that maintains heterochromatic regions such as centromeres. In contrast, in mammalian cells, gene silencing is known to occur through a post-transcriptional mechanism that uses exogenous short interfering RNAs or endogenous microRNAs to inactivate mRNA. We now show that the introduction of convergent transcription plasmids into either Schizosaccharomyces pombe or mammalian cells allows the production of double-stranded RNA from inserted gene fragments, resulting in TGS of endogenous genes. We predict that using convergent transcription to induce gene silencing will be a generally useful strategy and allow for a fuller molecular understanding of the biology of TGS.
Figures

Similar articles
-
A single Argonaute protein mediates both transcriptional and posttranscriptional silencing in Schizosaccharomyces pombe.Genes Dev. 2004 Oct 1;18(19):2359-67. doi: 10.1101/gad.1218004. Epub 2004 Sep 15. Genes Dev. 2004. PMID: 15371329 Free PMC article.
-
RNAi-mediated gene silencing in zebrafish triggered by convergent transcription.Sci Rep. 2014 Jun 9;4:5222. doi: 10.1038/srep05222. Sci Rep. 2014. PMID: 24909225 Free PMC article.
-
Spt6 is required for heterochromatic silencing in the fission yeast Schizosaccharomyces pombe.Mol Cell Biol. 2011 Oct;31(20):4193-204. doi: 10.1128/MCB.05568-11. Epub 2011 Aug 15. Mol Cell Biol. 2011. PMID: 21844224 Free PMC article.
-
Transcriptional gene silencing by short interfering RNAs.Curr Opin Mol Ther. 2005 Apr;7(2):125-31. Curr Opin Mol Ther. 2005. PMID: 15844619 Review.
-
RNA and epigenetic silencing: insight from fission yeast.Dev Growth Differ. 2012 Jan;54(1):129-41. doi: 10.1111/j.1440-169X.2011.01310.x. Epub 2011 Dec 12. Dev Growth Differ. 2012. PMID: 22150237 Free PMC article. Review.
Cited by
-
Characterization of novel transcripts in pseudorabies virus.Viruses. 2015 May 22;7(5):2727-44. doi: 10.3390/v7052727. Viruses. 2015. PMID: 26008709 Free PMC article.
-
Transpositional shuffling and quality control in male germ cells to enhance evolution of complex organisms.Ann N Y Acad Sci. 2015 Apr;1341(1):156-63. doi: 10.1111/nyas.12608. Epub 2014 Dec 31. Ann N Y Acad Sci. 2015. PMID: 25557795 Free PMC article.
-
Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution.Cell. 2015 Apr 23;161(3):541-554. doi: 10.1016/j.cell.2015.03.010. Cell. 2015. PMID: 25910208 Free PMC article.
-
Head-on and co-directional RNA polymerase collisions orchestrate bidirectional transcription termination.Mol Cell. 2023 Apr 6;83(7):1153-1164.e4. doi: 10.1016/j.molcel.2023.02.017. Epub 2023 Mar 13. Mol Cell. 2023. PMID: 36917983 Free PMC article.
-
Identification of cell-type specific alternative transcripts in the multicellular alga Volvox carteri.BMC Genomics. 2023 Oct 30;24(1):654. doi: 10.1186/s12864-023-09558-0. BMC Genomics. 2023. PMID: 37904088 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

