Nuclear domain 10-associated proteins recognize and segregate intranuclear DNA/protein complexes to negate gene expression
- PMID: 23021128
- PMCID: PMC3502357
- DOI: 10.1186/1743-422X-9-222
Nuclear domain 10-associated proteins recognize and segregate intranuclear DNA/protein complexes to negate gene expression
Abstract
Background: DNA viruses, such as herpes simplex virus type 1 (HSV-1), Simian virus 40 (SV40), and Cytomegaloviruses (CMV), start their replicative processes and transcription at specific nuclear domains known as ND10 (nuclear domain 10, also called PML bodies). It has been previously determined that for HSV-1 and SV40, a short DNA sequence and its binding protein are required and sufficient for cell localization of viral DNA replication and gene transcription.
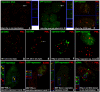
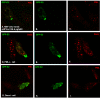
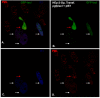
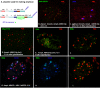
Results: Our recent observations provide evidence that a foreign (not endogenous) DNA/protein complex in the nucleus recruits ND10 proteins. First, the complexes formed from the bacterial lac operator DNA and its binding protein (lac repressor), or from HPV11 (human papillomavirus 11) origin DNA and its binding protein (E2), co-localized with different ND10 proteins. Second, the HSV-1 amplicon without inserted lac operator DNA repeats distributed in the nucleus randomly, whereas the amplicon with lac operator DNA repeats associated with ND10, suggesting that DNA-binding proteins are required to localize at ND10. The cellular intrinsic DNA/protein complex (as detected for U2 DNA) showed no association with ND10. Furthermore, our examination of PML-/-, Daxx-/-, and Sp100-negative cells led to our discovering that DNA/protein complexes recruit ND10 protein independently. Using the GFP-LacI/Operator system, we were able to direct the transfected DNA to ND10 and found that gene expression was significantly repressed when the transfected DNA was directed to ND10.
Conclusion: Taken together, the results suggest that cells recognize DNA/protein complexes through a mechanism that involves interaction with the ND10-associated proteins.
Figures
Similar articles
-
Mechanisms of Host IFI16, PML, and Daxx Protein Restriction of Herpes Simplex Virus 1 Replication.J Virol. 2018 Apr 27;92(10):e00057-18. doi: 10.1128/JVI.00057-18. Print 2018 May 15. J Virol. 2018. PMID: 29491153 Free PMC article.
-
Determination of minimum herpes simplex virus type 1 components necessary to localize transcriptionally active DNA to ND10.J Virol. 2003 May;77(10):5821-8. doi: 10.1128/jvi.77.10.5821-5828.2003. J Virol. 2003. PMID: 12719575 Free PMC article.
-
Effect of SUMO-SIM Interaction on the ICP0-Mediated Degradation of PML Isoform II and Its Associated Proteins in Herpes Simplex Virus 1 Infection.J Virol. 2020 Jun 1;94(12):e00470-20. doi: 10.1128/JVI.00470-20. Print 2020 Jun 1. J Virol. 2020. PMID: 32295906 Free PMC article.
-
Role of ND10 nuclear bodies in the chromatin repression of HSV-1.Virol J. 2016 Apr 5;13:62. doi: 10.1186/s12985-016-0516-4. Virol J. 2016. PMID: 27048561 Free PMC article. Review.
-
The Role of ND10 Nuclear Bodies in Herpesvirus Infection: A Frenemy for the Virus?Viruses. 2021 Feb 3;13(2):239. doi: 10.3390/v13020239. Viruses. 2021. PMID: 33546431 Free PMC article. Review.
Cited by
-
The PML-Interacting Protein DAXX: Histone Loading Gets into the Picture.Front Oncol. 2013 Jun 7;3:152. doi: 10.3389/fonc.2013.00152. eCollection 2013. Front Oncol. 2013. PMID: 23760585 Free PMC article.
-
Viral miRNA regulation of host gene expression.Semin Cell Dev Biol. 2023 Sep 15;146:2-19. doi: 10.1016/j.semcdb.2022.11.007. Epub 2022 Nov 30. Semin Cell Dev Biol. 2023. PMID: 36463091 Free PMC article. Review.
-
PML nuclear body-residing proteins sequentially associate with HPV genome after infectious nuclear delivery.PLoS Pathog. 2019 Feb 25;15(2):e1007590. doi: 10.1371/journal.ppat.1007590. eCollection 2019 Feb. PLoS Pathog. 2019. PMID: 30802273 Free PMC article.
-
Control of Viral Latency by Episome Maintenance Proteins.Trends Microbiol. 2020 Feb;28(2):150-162. doi: 10.1016/j.tim.2019.09.002. Epub 2019 Oct 14. Trends Microbiol. 2020. PMID: 31624007 Free PMC article. Review.
-
The Role of Promyelocytic Leukemia Nuclear Bodies During HPV Infection.Front Cell Infect Microbiol. 2020 Feb 19;10:35. doi: 10.3389/fcimb.2020.00035. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32154186 Free PMC article. Review.
References
-
- Spector DL. SnapShot: cellular bodies. Cell. 2006;127:1071. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

