Comprehensive analysis of host cellular interactions with human papillomavirus E6 proteins identifies new E6 binding partners and reflects viral diversity
- PMID: 23015706
- PMCID: PMC3503137
- DOI: 10.1128/JVI.02172-12
Comprehensive analysis of host cellular interactions with human papillomavirus E6 proteins identifies new E6 binding partners and reflects viral diversity
Abstract
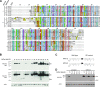
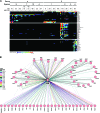
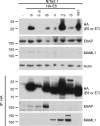
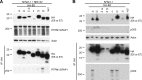
We have begun to define the human papillomavirus (HPV)-associated proteome for a subset of the more than 120 HPV types that have been identified to date. Our approach uses a mass spectrometry-based platform for the systematic identification of interactions between human papillomavirus and host cellular proteins, and here we report a proteomic analysis of the E6 proteins from 16 different HPV types. The viruses included represent high-risk, low-risk, and non-cancer-associated types from genus alpha as well as viruses from four different species in genus beta. The E6 interaction data set consists of 153 cellular proteins, including several previously reported HPV E6 interactors such as p53, E6AP, MAML1, and p300/CBP and proteins containing PDZ domains. We report the genus-specific binding of E6s to either E6AP or MAML1, define the specific HPV E6s that bind to p300, and demonstrate several new features of interactions involving beta HPV E6s. In particular, we report that several beta HPV E6s bind to proteins containing PDZ domains and that at least two beta HPV E6s bind to p53. Finally, we report the newly discovered interaction of proteins of E6 of beta genus, species 2, with the Ccr4-Not complex, the first report of a viral protein binding to this complex. This data set represents a comprehensive survey of E6 binding partners that provides a resource for the HPV field and will allow continued studies on the diverse biology of the human papillomaviruses.
Figures




Similar articles
-
Association of papillomavirus E6 proteins with either MAML1 or E6AP clusters E6 proteins by structure, function, and evolutionary relatedness.PLoS Pathog. 2017 Dec 27;13(12):e1006781. doi: 10.1371/journal.ppat.1006781. eCollection 2017 Dec. PLoS Pathog. 2017. PMID: 29281732 Free PMC article.
-
MAML1-induced HPV E6 oncoprotein stability is required for cellular proliferation and migration of cervical tumor-derived cells.J Med Virol. 2023 Mar;95(3):e28624. doi: 10.1002/jmv.28624. J Med Virol. 2023. PMID: 36852660
-
The human papillomavirus type 16 E6 oncoprotein can down-regulate p53 activity by targeting the transcriptional coactivator CBP/p300.J Virol. 1999 Aug;73(8):6209-19. doi: 10.1128/JVI.73.8.6209-6219.1999. J Virol. 1999. PMID: 10400710 Free PMC article.
-
Proteomic approaches to the study of papillomavirus-host interactions.Virology. 2013 Jan 5;435(1):57-69. doi: 10.1016/j.virol.2012.09.046. Virology. 2013. PMID: 23217616 Free PMC article. Review.
-
Structure and function of the papillomavirus E6 protein and its interacting proteins.Front Biosci. 2008 Jan 1;13:121-34. doi: 10.2741/2664. Front Biosci. 2008. PMID: 17981532 Review.
Cited by
-
A human papillomavirus 16 E2-TopBP1 dependent SIRT1-p300 acetylation switch regulates mitotic viral and human protein levels and activates the DNA damage response.mBio. 2024 Jun 12;15(6):e0067624. doi: 10.1128/mbio.00676-24. Epub 2024 May 9. mBio. 2024. PMID: 38722185 Free PMC article.
-
Recent advances in the study of HPV-associated carcinogenesis.Virol Sin. 2015 Apr;30(2):101-6. doi: 10.1007/s12250-015-3586-3. Epub 2015 Apr 20. Virol Sin. 2015. PMID: 25910482 Free PMC article. Review.
-
The difference of transcriptome of HPV-infected patients contributes more to the occurrence of cervical cancer than the mutations of E6 and E7 genes in HPV16.Medicine (Baltimore). 2024 Jan 19;103(3):e36822. doi: 10.1097/MD.0000000000036822. Medicine (Baltimore). 2024. PMID: 38241590 Free PMC article.
-
Papillomavirus E6 oncoproteins.Virology. 2013 Oct;445(1-2):115-37. doi: 10.1016/j.virol.2013.04.026. Epub 2013 May 24. Virology. 2013. PMID: 23711382 Free PMC article. Review.
-
The SMC5/6 Complex Interacts with the Papillomavirus E2 Protein and Influences Maintenance of Viral Episomal DNA.J Virol. 2018 Jul 17;92(15):e00356-18. doi: 10.1128/JVI.00356-18. Print 2018 Aug 1. J Virol. 2018. PMID: 29848583 Free PMC article.
References
-
- Akgül B, Cooke JC, Storey A. 2006. HPV-associated skin disease. J. Pathol. 208:165–175 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

