Surveillance and genome analysis of human bocavirus in patients with respiratory infection in Guangzhou, China
- PMID: 22984581
- PMCID: PMC3439446
- DOI: 10.1371/journal.pone.0044876
Surveillance and genome analysis of human bocavirus in patients with respiratory infection in Guangzhou, China
Abstract
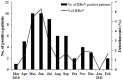
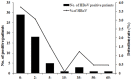
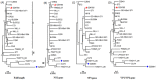
Human bocavirus (HBoV) is a novel parvovirus associated with respiratory tract diseases and gastrointestinal illness in adult and pediatric patients throughout the world. To investigate the epidemiological and genetic variation of HBoV in Guangzhou, South China, we screened 3460 throat swab samples from 1686 children and 1774 adults with acute respiratory infection symptoms for HBoV between March 2010 and February 2011, and analyzed the complete genome sequence of 2 HBoV strains. Specimens were screened for HBoV by real-time PCR and other 6 common respiratory viruses by RT-PCR or PCR. HBoV was detected in 58 (1.68%) out of 3460 samples, mostly from pediatric patients (52/58) and inpatient children (47/58). Six adult patients were detected as HBoV positive and 5 were emergency cases. Of these HBoV positive cases, 19 (32.76%) had co-pathogens including influenza virus (n = 5), RSV (n = 5), parainfluenza (n = 4), adenovirus (n = 1), coronavirus (n = 7). The complete genome sequences of 2 HBoVs strains (Genbank no. JN794565 and JN794566) were analyzed. Phylogenetic analysis showed that the 2 HBoV strains were HBoV1, and were most genetically close to ST2 (GenBank accession number DQ0000496). Recombination analysis confirmed that HBoV strain GZ9081 was an intra-genotype recombinant strain among HBoV1 variants.
Conflict of interest statement
Figures

Similar articles
-
Genetic characterization of human bocavirus among children with severe acute respiratory infection in China.J Infect. 2016 Aug;73(2):155-63. doi: 10.1016/j.jinf.2016.05.014. Epub 2016 Jun 13. J Infect. 2016. PMID: 27306487 Free PMC article.
-
Detection of human bocavirus from children and adults with acute respiratory tract illness in Guangzhou, southern China.BMC Infect Dis. 2011 Dec 14;11:345. doi: 10.1186/1471-2334-11-345. BMC Infect Dis. 2011. PMID: 22168387 Free PMC article.
-
Human bocavirus infection in young children with acute respiratory tract infection in Lanzhou, China.J Med Virol. 2010 Feb;82(2):282-8. doi: 10.1002/jmv.21689. J Med Virol. 2010. PMID: 20029808 Free PMC article.
-
Epidemic and molecular evolution of human bocavirus in hospitalized children with acute respiratory tract infection.Eur J Clin Microbiol Infect Dis. 2015 Jan;34(1):75-81. doi: 10.1007/s10096-014-2215-7. Epub 2014 Jul 29. Eur J Clin Microbiol Infect Dis. 2015. PMID: 25070494 Free PMC article.
-
Human bocavirus: Current knowledge and future challenges.World J Gastroenterol. 2016 Oct 21;22(39):8684-8697. doi: 10.3748/wjg.v22.i39.8684. World J Gastroenterol. 2016. PMID: 27818586 Free PMC article. Review.
Cited by
-
Detection of bocavirus in children suffering from acute respiratory tract infections in Saudi Arabia.PLoS One. 2013;8(1):e55500. doi: 10.1371/journal.pone.0055500. Epub 2013 Jan 30. PLoS One. 2013. PMID: 23383205 Free PMC article.
-
Complete genomes of three human bocavirus strains from children with gastroenteritis and respiratory tract illnesses in Jiangsu, China.J Virol. 2012 Dec;86(24):13826-7. doi: 10.1128/JVI.02618-12. J Virol. 2012. PMID: 23166240 Free PMC article.
-
Genetic variability of human metapneumo- and bocaviruses in children with respiratory tract infections.Influenza Other Respir Viruses. 2014 Jan;8(1):107-15. doi: 10.1111/irv.12185. Epub 2013 Nov 7. Influenza Other Respir Viruses. 2014. PMID: 24373295 Free PMC article.
-
The spectrum of viral pathogens in children with severe acute lower respiratory tract infection: A 3-year prospective study in the pediatric intensive care unit.J Med Virol. 2019 Sep;91(9):1633-1642. doi: 10.1002/jmv.25502. Epub 2019 Jun 13. J Med Virol. 2019. PMID: 31081548 Free PMC article.
-
Genetic characterization of human bocavirus among children with severe acute respiratory infection in China.J Infect. 2016 Aug;73(2):155-63. doi: 10.1016/j.jinf.2016.05.014. Epub 2016 Jun 13. J Infect. 2016. PMID: 27306487 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

