Evolution of neurodegeneration
- PMID: 22975006
- PMCID: PMC4662249
- DOI: 10.1016/j.cub.2012.07.008
Evolution of neurodegeneration
Abstract
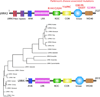
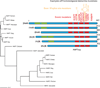
A number of neurodegenerative diseases principally affect humans as they age and are characterized by the loss of specific groups of neurons in different brain regions. Although these disorders are generally sporadic, it is now clear that many of them have a substantial genetic component. As genes are the raw material with which evolution works, we might benefit from understanding these genes in an evolutionary framework. Here, I will discuss how we can understand whether evolution has shaped genes involved in neurodegeneration and the implications for practical issues, such as our choice of model systems for studying these diseases, and more theoretical concerns, such as the level of selection against these phenotypes.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures


Similar articles
-
Argyrophilic grain disease.Handb Clin Neurol. 2008;89:553-63. doi: 10.1016/S0072-9752(07)01251-1. Handb Clin Neurol. 2008. PMID: 18631777 Review. No abstract available.
-
Tau protein mutations confirmed as neuron killers.Science. 1998 Jun 5;280(5369):1524-5. doi: 10.1126/science.280.5369.1524. Science. 1998. PMID: 9644015 No abstract available.
-
Pathogenic implications of mutations in the tau gene in pallido-ponto-nigral degeneration and related neurodegenerative disorders linked to chromosome 17.Proc Natl Acad Sci U S A. 1998 Oct 27;95(22):13103-7. doi: 10.1073/pnas.95.22.13103. Proc Natl Acad Sci U S A. 1998. PMID: 9789048 Free PMC article.
-
Neuroepidemiologic research initiatives on Guam: past and present.Neuroepidemiology. 1999;18(6):279-91. doi: 10.1159/000026223. Neuroepidemiology. 1999. PMID: 10545780 Review.
-
ɑ-Synuclein strains and the variable pathologies of synucleinopathies.J Neurochem. 2016 Oct;139 Suppl 1:256-274. doi: 10.1111/jnc.13595. Epub 2016 Mar 30. J Neurochem. 2016. PMID: 26924014 Review.
Cited by
-
Did α-Synuclein and Glucocerebrosidase Coevolve? Implications for Parkinson's Disease.PLoS One. 2015 Jul 27;10(7):e0133863. doi: 10.1371/journal.pone.0133863. eCollection 2015. PLoS One. 2015. PMID: 26214314 Free PMC article.
-
An evolutionary roadmap to the microtubule-associated protein MAP Tau.BMC Genomics. 2016 Mar 31;17:264. doi: 10.1186/s12864-016-2590-9. BMC Genomics. 2016. PMID: 27030133 Free PMC article.
-
Chronic oxidative damage together with genome repair deficiency in the neurons is a double whammy for neurodegeneration: Is damage response signaling a potential therapeutic target?Mech Ageing Dev. 2017 Jan;161(Pt A):163-176. doi: 10.1016/j.mad.2016.09.005. Epub 2016 Sep 20. Mech Ageing Dev. 2017. PMID: 27663141 Free PMC article. Review.
-
Human-lineage-specific genomic elements are associated with neurodegenerative disease and APOE transcript usage.Nat Commun. 2021 Apr 6;12(1):2076. doi: 10.1038/s41467-021-22262-5. Nat Commun. 2021. PMID: 33824317 Free PMC article.
-
Hominin-specific regulatory elements selectively emerged in oligodendrocytes and are disrupted in autism patients.Nat Commun. 2020 Jan 16;11(1):301. doi: 10.1038/s41467-019-14269-w. Nat Commun. 2020. PMID: 31949148 Free PMC article.
References
-
- Hardy J, Orr H. The genetics of neurodegenerative diseases. J. Neurochem. 2006;97:1690–1699. - PubMed
-
- Lill CM, Bertram L. Towards unveiling the genetics of neurodegenerative diseases. Semin Neurol. 2011;31:531–541. - PubMed
-
- Halliday G, Lees A, Stern M. Milestones in Parkinson’s disease--clinical and pathologic features. Mov. Disord. 2011;26:1015–1021. - PubMed
-
- Shulman JM, De Jager PL, Feany MB. Parkinson’s disease: genetics and pathogenesis. Annu Rev Pathol. 2011;6:193–222. - PubMed
-
- Gasser T, Hardy J, Mizuno Y. Milestones in PD genetics. Mov. Disord. 2011;26:1042–1048. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

