Genomics and localization of the Arabidopsis DHHC-cysteine-rich domain S-acyltransferase protein family
- PMID: 22968831
- PMCID: PMC3490592
- DOI: 10.1104/pp.112.203968
Genomics and localization of the Arabidopsis DHHC-cysteine-rich domain S-acyltransferase protein family
Abstract
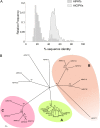
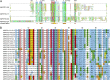
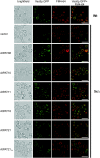
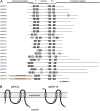
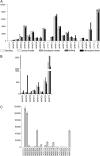
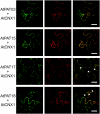
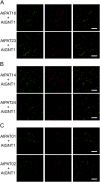
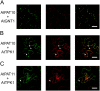
Protein lipid modification of cysteine residues, referred to as S-palmitoylation or S-acylation, is an important secondary and reversible modification that regulates membrane association, trafficking, and function of target proteins. This enzymatic reaction is mediated by protein S-acyl transferases (PATs). Here, the phylogeny, genomic organization, protein topology, expression, and localization pattern of the 24 PAT family members from Arabidopsis (Arabidopsis thaliana) is described. Most PATs are expressed at ubiquitous levels and tissues throughout the development, while few genes are expressed especially during flower development preferentially in pollen and stamen. The proteins display large sequence and structural variations but exhibit a common protein topology that is preserved in PATs from various organisms. Arabidopsis PAT proteins display a complex targeting pattern and were detected at the endoplasmic reticulum, Golgi, endosomal compartments, and the vacuolar membrane. However, most proteins were targeted to the plasma membrane. This large concentration of plant PAT activity to the plasma membrane suggests that the plant cellular S-acylation machinery is functionally different compared with that of yeast (Saccharomyces cerevisiae) and mammalians.
Figures
Similar articles
-
A Golgi and tonoplast localized S-acyl transferase is involved in cell expansion, cell division, vascular patterning and fertility in Arabidopsis.New Phytol. 2013 Oct;200(2):444-456. doi: 10.1111/nph.12385. Epub 2013 Jun 25. New Phytol. 2013. PMID: 23795888 Free PMC article.
-
Putative DHHC-cysteine-rich domain S-acyltransferase in plants.PLoS One. 2013 Oct 14;8(10):e75985. doi: 10.1371/journal.pone.0075985. eCollection 2013. PLoS One. 2013. PMID: 24155879 Free PMC article.
-
Both male and female gametogenesis require a fully functional protein S-acyl transferase 21 in Arabidopsis thaliana.Plant J. 2019 Nov;100(4):754-767. doi: 10.1111/tpj.14475. Epub 2019 Sep 12. Plant J. 2019. PMID: 31369173
-
Protein palmitoylation by a family of DHHC protein S-acyltransferases.J Lipid Res. 2006 Jun;47(6):1118-27. doi: 10.1194/jlr.R600007-JLR200. Epub 2006 Apr 1. J Lipid Res. 2006. PMID: 16582420 Review.
-
Structure and function of DHHC protein S-acyltransferases.Biochem Soc Trans. 2017 Aug 15;45(4):923-8. doi: 10.1042/BST20160304. Epub 2017 Jun 19. Biochem Soc Trans. 2017. PMID: 28630137 Review.
Cited by
-
Function of membrane domains in rho-of-plant signaling.Plant Physiol. 2021 Apr 2;185(3):663-681. doi: 10.1093/plphys/kiaa082. Plant Physiol. 2021. PMID: 33793925 Free PMC article. Review.
-
Progress toward Understanding Protein S-acylation: Prospective in Plants.Front Plant Sci. 2017 Mar 24;8:346. doi: 10.3389/fpls.2017.00346. eCollection 2017. Front Plant Sci. 2017. PMID: 28392791 Free PMC article. Review.
-
Genome-Wide Identification and Expression Pattern Profiling of the Aquaporin Gene Family in Papaya (Carica papaya L.).Int J Mol Sci. 2023 Dec 8;24(24):17276. doi: 10.3390/ijms242417276. Int J Mol Sci. 2023. PMID: 38139107 Free PMC article.
-
Oligomerization of DHHC protein S-acyltransferases.J Biol Chem. 2013 Aug 2;288(31):22862-70. doi: 10.1074/jbc.M113.458794. Epub 2013 Jun 22. J Biol Chem. 2013. PMID: 23793055 Free PMC article.
-
A Golgi and tonoplast localized S-acyl transferase is involved in cell expansion, cell division, vascular patterning and fertility in Arabidopsis.New Phytol. 2013 Oct;200(2):444-456. doi: 10.1111/nph.12385. Epub 2013 Jun 25. New Phytol. 2013. PMID: 23795888 Free PMC article.
References
-
- Adjobo-Hermans MJ, Goedhart J, Gadella TW., Jr (2006) Plant G protein heterotrimers require dual lipidation motifs of Gα and Gγ and do not dissociate upon activation. J Cell Sci 119: 5087–5097 - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 - PubMed
-
- Baekkeskov S, Kanaani J. (2009) Palmitoylation cycles and regulation of protein function (Review). Mol Membr Biol 26: 42–54 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

