Modulation of inflammatory gene expression by the ribotoxin deoxynivalenol involves coordinate regulation of the transcriptome and translatome
- PMID: 22968694
- PMCID: PMC3537125
- DOI: 10.1093/toxsci/kfs266
Modulation of inflammatory gene expression by the ribotoxin deoxynivalenol involves coordinate regulation of the transcriptome and translatome
Abstract
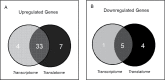
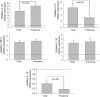
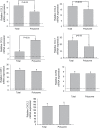
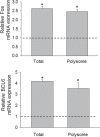
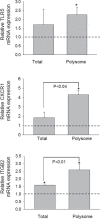
The trichothecene deoxynivalenol (DON), a common contaminant of cereal-based foods, is a ribotoxic mycotoxin known to activate innate immune cells in vivo and in vitro. Although it is recognized that DON induces transcription and mRNA stabilization of inflammation-associated mRNAs in mononuclear phagocytes, it is not known if this toxin affects translation of selected mRNA species in the cellular pool. To address this question, we employed a focused inflammation/autoimmunity PCR array to compare DON-induced changes in profiles of polysome-associated mRNA transcripts (translatome) to total cellular mRNA transcripts (transcriptome) in the RAW 264.7 murine macrophage model. Exposure to DON at 250 ng/ml (0.84 µM) for 6 h induced robust expression changes in inflammatory response genes including cytokines, cytokine receptors, chemokines, chemokine receptors, and transcription factors, with 73% of the changes being highly comparable within transcriptome and translatome populations. When expression changes of selected representative inflammatory response genes in the polysome and cellular mRNA pools were quantified in a follow-up study by real-time PCR, closely coordinated regulation of the translatome and transcriptome was confirmed; however, modest but significant differences in the relative expression of some genes within the two pools were also detectable. Taken together, DON's capacity to alter translation expression of inflammation-associated genes appears to be driven predominantly by selective transcription and mRNA stabilization that have been reported previously; however, a small subset of these genes appear to be further regulated at the translational level.
Figures


Similar articles
-
Dynamic changes in ribosome-associated proteome and phosphoproteome during deoxynivalenol-induced translation inhibition and ribotoxic stress.Toxicol Sci. 2014 Mar;138(1):217-33. doi: 10.1093/toxsci/kft270. Epub 2013 Nov 27. Toxicol Sci. 2014. PMID: 24284785 Free PMC article.
-
Effects of oral exposure to naturally-occurring and synthetic deoxynivalenol congeners on proinflammatory cytokine and chemokine mRNA expression in the mouse.Toxicol Appl Pharmacol. 2014 Jul 15;278(2):107-15. doi: 10.1016/j.taap.2014.04.016. Epub 2014 Apr 29. Toxicol Appl Pharmacol. 2014. PMID: 24793808 Free PMC article.
-
Gene expression profiling in spleens of deoxynivalenol-exposed mice: immediate early genes as primary targets.J Toxicol Environ Health A. 2004 Sep 24;67(18):1423-41. doi: 10.1080/15287390490483827. J Toxicol Environ Health A. 2004. PMID: 15371230
-
Deoxynivalenol-induced proinflammatory gene expression: mechanisms and pathological sequelae.Toxins (Basel). 2010 Jun;2(6):1300-17. doi: 10.3390/toxins2061300. Epub 2010 Jun 1. Toxins (Basel). 2010. PMID: 22069639 Free PMC article. Review.
-
Toxicology of deoxynivalenol (vomitoxin).J Toxicol Environ Health. 1996 May;48(1):1-34. doi: 10.1080/009841096161447. J Toxicol Environ Health. 1996. PMID: 8637056 Review.
Cited by
-
The food contaminant deoxynivalenol provokes metabolic impairments resulting in non-alcoholic fatty liver (NAFL) in mice.Sci Rep. 2020 Jul 21;10(1):12072. doi: 10.1038/s41598-020-68712-w. Sci Rep. 2020. PMID: 32694515 Free PMC article.
-
Deoxynivalenol enhances IL-1ß expression in BV2 microglial cells through activation of the NF-?B pathway and the ASC/NLRP3 inflammasome.EXCLI J. 2019 Jun 11;18:356-369. doi: 10.17179/excli2018-1974. eCollection 2019. EXCLI J. 2019. PMID: 31338007 Free PMC article.
-
Dynamic changes in ribosome-associated proteome and phosphoproteome during deoxynivalenol-induced translation inhibition and ribotoxic stress.Toxicol Sci. 2014 Mar;138(1):217-33. doi: 10.1093/toxsci/kft270. Epub 2013 Nov 27. Toxicol Sci. 2014. PMID: 24284785 Free PMC article.
-
Effects of oral exposure to naturally-occurring and synthetic deoxynivalenol congeners on proinflammatory cytokine and chemokine mRNA expression in the mouse.Toxicol Appl Pharmacol. 2014 Jul 15;278(2):107-15. doi: 10.1016/j.taap.2014.04.016. Epub 2014 Apr 29. Toxicol Appl Pharmacol. 2014. PMID: 24793808 Free PMC article.
-
Deoxynivalenol affects cell metabolism in vivo and inhibits protein synthesis in IPEC-1 cells.Mycotoxin Res. 2023 Aug;39(3):219-231. doi: 10.1007/s12550-023-00489-z. Epub 2023 May 31. Mycotoxin Res. 2023. PMID: 37256505 Free PMC article.

