The Wnt/β-catenin/T-cell factor 4 pathway up-regulates high-mobility group A1 expression in colon cancer
- PMID: 22961697
- PMCID: PMC3616152
- DOI: 10.1002/cbf.2876
The Wnt/β-catenin/T-cell factor 4 pathway up-regulates high-mobility group A1 expression in colon cancer
Abstract
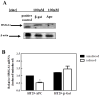
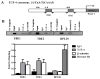
High-mobility group A1 (HMGA1) encodes proteins that act as mediators in viral integration, modification of chromatin structure, neoplastic transformation and metastatic progression. Because HMGA1 is overexpressed in most cancers and has transcriptional relationships with several Wnt-responsive genes, we explored the involvement of HMGA1 in Wnt/β-catenin/TCF-4 signalling. In adenomatous polyposis coli (APC(Min/+)) mice, we observed significant up-regulation of HMGA1 mRNA and protein in intestinal tumours when compared with normal intestinal mucosa. Conversely, restoration of Wnt signalling by the zinc induction of wild-type APC resulted in HMGA1 down-regulation in HT-29 cells. Because APC mutations are associated with mobilization of the β-catenin/TCF-4 transcriptional complex and subsequent activation of downstream oncogenic targets, we analyzed the 5'-flanking sequence of HMGA1 for putative TCF-4 binding elements. We identified two regions that specifically bind the β-catenin/TCF-4 complex in vitro and in vivo, identifying HMGA1 as an immediate target of the β-catenin/TCF-4 signalling pathway in colon cancer. Collectively, these findings strongly implicate Wnt/β-catenin/TCF-4 signalling in regulating HMGA1 to further expand the extensive regulatory network affected by Wnt/β-catenin/TCF-4 signalling.
Copyright © 2012 John Wiley & Sons, Ltd.
Figures


Similar articles
-
Vitamin D receptor deficiency enhances Wnt/β-catenin signaling and tumor burden in colon cancer.PLoS One. 2011;6(8):e23524. doi: 10.1371/journal.pone.0023524. Epub 2011 Aug 15. PLoS One. 2011. PMID: 21858154 Free PMC article.
-
Regulation of cyclooxygenase-2 expression by the Wnt and ras pathways.Cancer Res. 2003 Feb 1;63(3):728-34. Cancer Res. 2003. PMID: 12566320
-
Neurotensin receptor 1 gene activation by the Tcf/beta-catenin pathway is an early event in human colonic adenomas.Carcinogenesis. 2006 Apr;27(4):708-16. doi: 10.1093/carcin/bgi269. Epub 2005 Nov 19. Carcinogenesis. 2006. PMID: 16299383
-
Transcriptional Regulation of Wnt/β-Catenin Pathway in Colorectal Cancer.Cells. 2020 Sep 19;9(9):2125. doi: 10.3390/cells9092125. Cells. 2020. PMID: 32961708 Free PMC article. Review.
-
The Yin-Yang of TCF/beta-catenin signaling.Adv Cancer Res. 2000;77:1-24. doi: 10.1016/s0065-230x(08)60783-6. Adv Cancer Res. 2000. PMID: 10549354 Review.
Cited by
-
Design, synthesis and evaluation of 4,7-disubstituted 8-methoxyquinazoline derivatives as potential cytotoxic agents targeting β-catenin/TCF4 signaling pathway.Transl Oncol. 2022 May;19:101395. doi: 10.1016/j.tranon.2022.101395. Epub 2022 Mar 21. Transl Oncol. 2022. PMID: 35325837 Free PMC article.
-
High mobility group a proteins as tumor markers.Front Med (Lausanne). 2015 Mar 25;2:15. doi: 10.3389/fmed.2015.00015. eCollection 2015. Front Med (Lausanne). 2015. PMID: 25859543 Free PMC article. Review.
-
The canonical Wnt/β-catenin signaling pathway stimulates herpes simplex virus 1 productive infection.Virus Res. 2018 Sep 2;256:29-37. doi: 10.1016/j.virusres.2018.07.020. Epub 2018 Aug 2. Virus Res. 2018. PMID: 30077727 Free PMC article.
-
Lessons from the Crypt: HMGA1-Amping up Wnt for Stem Cells and Tumor Progression.Cancer Res. 2018 Apr 15;78(8):1890-1897. doi: 10.1158/0008-5472.CAN-17-3045. Epub 2018 Apr 4. Cancer Res. 2018. PMID: 29618461 Free PMC article. Review.
-
The high mobility group A1 molecular switch: turning on cancer - can we turn it off?Expert Opin Ther Targets. 2014 May;18(5):541-53. doi: 10.1517/14728222.2014.900045. Epub 2014 Mar 31. Expert Opin Ther Targets. 2014. PMID: 24684280 Free PMC article. Review.
References
-
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. - PubMed
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. - PubMed
-
- Fulda S. Tumor resistance to apoptosis. Int J Cancer. 2009;124(3):511–5. - PubMed
-
- Blagosklonny MV. Molecular theory of cancer. Cancer Biol Ther. 2005;4(6):621–7. - PubMed
-
- Finlay GJ. Genetics, molecular biology and colorectal cancer. Mutat Res. 1993;290(1):3–12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

