Multi-level regulation of cellular recognition of viral dsRNA
- PMID: 22960755
- PMCID: PMC7079809
- DOI: 10.1007/s00018-012-1149-4
Multi-level regulation of cellular recognition of viral dsRNA
Abstract
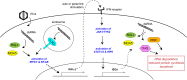
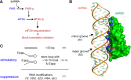
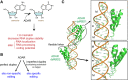
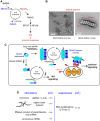
Effective antiviral immunity depends on accurate recognition of viral RNAs by the innate immune system. Double-stranded RNA (dsRNA) often accumulates in virally infected cells and was initially considered a unique viral signature that was sufficient to initiate antiviral response through dsRNA receptors and dsRNA-dependent effectors such as Toll-like receptor 3, retinoic acid inducible gene-1, protein kinase RNA-activated and oligoadenylate synthetase. However, dsRNA is also present in many cellular RNAs, raising a question of how these receptors and effectors discriminate between viral and cellular dsRNAs. Accumulating evidence suggests that innate immune sensors detect not only dsRNA structure but also other and often multiple features of RNA such as length, sequence, cellular location, post-transcriptional processing and modification, which are divergent between viral and cellular RNAs. This review summarizes recent findings on the substrate specificities of a few selected dsRNA-dependent effectors and receptors, which have revealed more complex mechanisms involved in cellular discrimination between self and non-self RNA.
Figures



Similar articles
-
Dissecting the roles of TRBP and PACT in double-stranded RNA recognition and processing of noncoding RNAs.Wiley Interdiscip Rev RNA. 2015 May-Jun;6(3):271-89. doi: 10.1002/wrna.1272. Epub 2015 Jan 28. Wiley Interdiscip Rev RNA. 2015. PMID: 25630541 Free PMC article. Review.
-
RIGorous detection: exposing virus through RNA sensing.Science. 2010 Jan 15;327(5963):284-6. doi: 10.1126/science.1185068. Science. 2010. PMID: 20075242
-
Kinetic mechanism for viral dsRNA length discrimination by MDA5 filaments.Proc Natl Acad Sci U S A. 2012 Dec 4;109(49):E3340-9. doi: 10.1073/pnas.1208618109. Epub 2012 Nov 5. Proc Natl Acad Sci U S A. 2012. PMID: 23129641 Free PMC article.
-
Processing of double-stranded RNA in mammalian cells: a direct antiviral role?J Interferon Cytokine Res. 2014 Jun;34(6):469-77. doi: 10.1089/jir.2014.0003. J Interferon Cytokine Res. 2014. PMID: 24905204 Review.
-
Double-Stranded RNA Sensors and Modulators in Innate Immunity.Annu Rev Immunol. 2019 Apr 26;37:349-375. doi: 10.1146/annurev-immunol-042718-041356. Epub 2019 Jan 23. Annu Rev Immunol. 2019. PMID: 30673536 Free PMC article. Review.
Cited by
-
Spiropyran as a potential molecular diagnostic tool for double-stranded RNA detection.BMC Biomed Eng. 2019 Mar 18;1:6. doi: 10.1186/s42490-019-0008-x. eCollection 2019. BMC Biomed Eng. 2019. PMID: 32903305 Free PMC article.
-
The N-Terminal α-Helix of Potato Virus X-Encoded RNA-Dependent RNA Polymerase Is Required for Membrane Association and Multimerization.Viruses. 2022 Aug 28;14(9):1907. doi: 10.3390/v14091907. Viruses. 2022. PMID: 36146714 Free PMC article.
-
Unconventional RNA-binding proteins step into the virus-host battlefront.Wiley Interdiscip Rev RNA. 2018 Nov;9(6):e1498. doi: 10.1002/wrna.1498. Epub 2018 Aug 9. Wiley Interdiscip Rev RNA. 2018. PMID: 30091184 Free PMC article. Review.
-
Double-Stranded-RNA-Binding Protein 2 Participates in Antiviral Defense.J Virol. 2020 May 18;94(11):e00017-20. doi: 10.1128/JVI.00017-20. Print 2020 May 18. J Virol. 2020. PMID: 32213615 Free PMC article.
-
SLIRP promotes autoimmune diseases by amplifying antiviral signaling via positive feedback regulation.bioRxiv [Preprint]. 2024 Jun 10:2024.03.28.587146. doi: 10.1101/2024.03.28.587146. bioRxiv. 2024. PMID: 38915695 Free PMC article. Preprint.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

