The great unravelling: chromatin as a modulator of the aging process
- PMID: 22959736
- PMCID: PMC3482262
- DOI: 10.1016/j.tibs.2012.08.001
The great unravelling: chromatin as a modulator of the aging process
Abstract
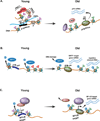
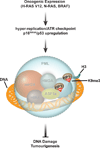
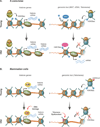
During embryogenesis, the establishment of chromatin states permits the implementation of genetic programs that allow the faithful development of the organism. However, these states are not fixed and there is much evidence that stochastic or chronic deterioration of chromatin organization, as correlated by transcriptional alterations and the accumulation of DNA damage in cells, occurs during the lifespan of the individual. Whether causal or simply a byproduct of macromolecular decay, these changes in chromatin states have emerged as potentially central conduits of mammalian aging. This review explores the current state of our understanding of the links between chromatin organization and aging.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures

Similar articles
-
Epigenetic Regulation of Chromatin States in Schizosaccharomyces pombe.Cold Spring Harb Perspect Biol. 2015 Jul 1;7(7):a018770. doi: 10.1101/cshperspect.a018770. Cold Spring Harb Perspect Biol. 2015. PMID: 26134317 Free PMC article. Review.
-
Small changes, big effects: chromatin goes aging.Subcell Biochem. 2013;61:151-76. doi: 10.1007/978-94-007-4525-4_8. Subcell Biochem. 2013. PMID: 23150251 Review.
-
The H1 linker histones: multifunctional proteins beyond the nucleosomal core particle.EMBO Rep. 2015 Nov;16(11):1439-53. doi: 10.15252/embr.201540749. Epub 2015 Oct 15. EMBO Rep. 2015. PMID: 26474902 Free PMC article. Review.
-
Replication timing maintains the global epigenetic state in human cells.Science. 2021 Apr 23;372(6540):371-378. doi: 10.1126/science.aba5545. Epub 2021 Apr 22. Science. 2021. PMID: 33888635 Free PMC article.
-
Centromeric heterochromatin assembly in fission yeast--balancing transcription, RNA interference and chromatin modification.Chromosome Res. 2012 Jul;20(5):521-34. doi: 10.1007/s10577-012-9288-x. Chromosome Res. 2012. PMID: 22733402 Free PMC article. Review.
Cited by
-
Functional implications of genome topology.Nat Struct Mol Biol. 2013 Mar;20(3):290-9. doi: 10.1038/nsmb.2474. Nat Struct Mol Biol. 2013. PMID: 23463314 Free PMC article. Review.
-
Toxic Y chromosome: Increased repeat expression and age-associated heterochromatin loss in male Drosophila with a young Y chromosome.PLoS Genet. 2021 Apr 22;17(4):e1009438. doi: 10.1371/journal.pgen.1009438. eCollection 2021 Apr. PLoS Genet. 2021. PMID: 33886541 Free PMC article.
-
Probing the depths of cellular senescence.J Cell Biol. 2013 Jul 8;202(1):11-3. doi: 10.1083/jcb.201305155. Epub 2013 Jul 1. J Cell Biol. 2013. PMID: 23816622 Free PMC article.
-
Immune senescence, epigenetics and autoimmunity.Clin Immunol. 2018 Nov;196:59-63. doi: 10.1016/j.clim.2018.04.002. Epub 2018 Apr 11. Clin Immunol. 2018. PMID: 29654845 Free PMC article. Review.
-
A role for SUV39H1-mediated H3K9 trimethylation in the control of genome stability and senescence in WI38 human diploid lung fibroblasts.Aging (Albany NY). 2014 Jul;6(7):545-63. doi: 10.18632/aging.100678. Aging (Albany NY). 2014. PMID: 25063769 Free PMC article.
References
-
- Herskind AM, et al. The heritability of human longevity: a population-based study of 2872 Danish twin pairs born 1870–1900. Hum. Genet. 1996;97:319–323. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

