LIN28 binds messenger RNAs at GGAGA motifs and regulates splicing factor abundance
- PMID: 22959275
- PMCID: PMC3483422
- DOI: 10.1016/j.molcel.2012.08.004
LIN28 binds messenger RNAs at GGAGA motifs and regulates splicing factor abundance
Abstract
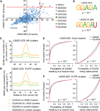
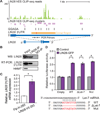
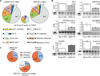
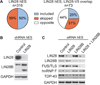
LIN28 is a conserved RNA-binding protein implicated in pluripotency, reprogramming, and oncogenesis. It was previously shown to act primarily by blocking let-7 microRNA (miRNA) biogenesis, but here we elucidate distinct roles of LIN28 regulation via its direct messenger RNA (mRNA) targets. Through crosslinking and immunoprecipitation coupled with high-throughput sequencing (CLIP-seq) in human embryonic stem cells and somatic cells expressing exogenous LIN28, we have defined discrete LIN28-binding sites in a quarter of human transcripts. These sites revealed that LIN28 binds to GGAGA sequences enriched within loop structures in mRNAs, reminiscent of its interaction with let-7 miRNA precursors. Among LIN28 mRNA targets, we found evidence for LIN28 autoregulation and also direct but differing effects on the protein abundance of splicing regulators in somatic and pluripotent stem cells. Splicing-sensitive microarrays demonstrated that exogenous LIN28 expression causes widespread downstream alternative splicing changes. These findings identify important regulatory functions of LIN28 via direct mRNA interactions.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Mechanisms of Lin28-mediated miRNA and mRNA regulation--a structural and functional perspective.Int J Mol Sci. 2013 Aug 9;14(8):16532-53. doi: 10.3390/ijms140816532. Int J Mol Sci. 2013. PMID: 23939427 Free PMC article. Review.
-
Identification of mRNAs bound and regulated by human LIN28 proteins and molecular requirements for RNA recognition.RNA. 2013 May;19(5):613-26. doi: 10.1261/rna.036491.112. Epub 2013 Mar 12. RNA. 2013. PMID: 23481595 Free PMC article.
-
Drosha mediates destabilization of Lin28 mRNA targets.Cell Cycle. 2012 Oct 1;11(19):3590-8. doi: 10.4161/cc.21871. Epub 2012 Aug 30. Cell Cycle. 2012. PMID: 22935707 Free PMC article.
-
The RNA-binding landscape of RBM10 and its role in alternative splicing regulation in models of mouse early development.RNA Biol. 2017 Jan 2;14(1):45-57. doi: 10.1080/15476286.2016.1247148. Epub 2016 Oct 20. RNA Biol. 2017. PMID: 27763814 Free PMC article.
-
Embryonic stem cell microRNAs: defining factors in induced pluripotent (iPS) and cancer (CSC) stem cells?Curr Stem Cell Res Ther. 2009 Sep;4(3):168-77. doi: 10.2174/157488809789057400. Curr Stem Cell Res Ther. 2009. PMID: 19492978 Review.
Cited by
-
RBPmotif: a web server for the discovery of sequence and structure preferences of RNA-binding proteins.Nucleic Acids Res. 2013 Jul;41(Web Server issue):W180-6. doi: 10.1093/nar/gkt463. Epub 2013 Jun 10. Nucleic Acids Res. 2013. PMID: 23754853 Free PMC article.
-
LOC101929709 promotes gastric cancer progression by aiding LIN28B to stabilize c-MYC mRNA.Gastric Cancer. 2023 Mar;26(2):169-186. doi: 10.1007/s10120-022-01348-z. Epub 2022 Oct 25. Gastric Cancer. 2023. PMID: 36284068
-
Lin28 Regulates Cancer Cell Stemness for Tumour Progression.Cancers (Basel). 2022 Sep 24;14(19):4640. doi: 10.3390/cancers14194640. Cancers (Basel). 2022. PMID: 36230562 Free PMC article. Review.
-
Resveratrol inhibits Lin28A expression and induces its degradation via the proteasomal pathway in NCCIT cells.Oncol Lett. 2024 Sep 30;28(6):577. doi: 10.3892/ol.2024.14710. eCollection 2024 Dec. Oncol Lett. 2024. PMID: 39397804 Free PMC article.
-
Imp interacts with Lin28 to regulate adult stem cell proliferation in the Drosophila intestine.PLoS Genet. 2022 Sep 7;18(9):e1010385. doi: 10.1371/journal.pgen.1010385. eCollection 2022 Sep. PLoS Genet. 2022. PMID: 36070313 Free PMC article.
References
-
- Balzer E, Heine C, Jiang Q, Lee VM, Moss EG. LIN28 alters cell fate succession and acts independently of the let-7 microRNA during neurogliogenesis in vitro. Development. 2010;137:891–900. - PubMed
-
- Balzer E, Moss EG. Localization of the developmental timing regulator Lin28 to mRNP complexes, P-bodies and stress granules. RNA Biol. 2007;4:16–25. - PubMed
-
- Bernhart SH, Hofacker IL, Stadler PF. Local RNA base pairing probabilities in large sequences. Bioinformatics. 2006;22:614–615. - PubMed
-
- Bussing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med. 2008;14:400–409. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

