An expansive human regulatory lexicon encoded in transcription factor footprints
- PMID: 22955618
- PMCID: PMC3736582
- DOI: 10.1038/nature11212
An expansive human regulatory lexicon encoded in transcription factor footprints
Abstract
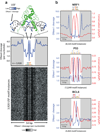
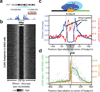
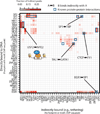
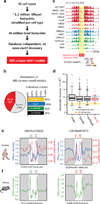
Regulatory factor binding to genomic DNA protects the underlying sequence from cleavage by DNase I, leaving nucleotide-resolution footprints. Using genomic DNase I footprinting across 41 diverse cell and tissue types, we detected 45 million transcription factor occupancy events within regulatory regions, representing differential binding to 8.4 million distinct short sequence elements. Here we show that this small genomic sequence compartment, roughly twice the size of the exome, encodes an expansive repertoire of conserved recognition sequences for DNA-binding proteins that nearly doubles the size of the human cis-regulatory lexicon. We find that genetic variants affecting allelic chromatin states are concentrated in footprints, and that these elements are preferentially sheltered from DNA methylation. High-resolution DNase I cleavage patterns mirror nucleotide-level evolutionary conservation and track the crystallographic topography of protein-DNA interfaces, indicating that transcription factor structure has been evolutionarily imprinted on the human genome sequence. We identify a stereotyped 50-base-pair footprint that precisely defines the site of transcript origination within thousands of human promoters. Finally, we describe a large collection of novel regulatory factor recognition motifs that are highly conserved in both sequence and function, and exhibit cell-selective occupancy patterns that closely parallel major regulators of development, differentiation and pluripotency.
Figures


Comment in
-
Genomics: users' guide to the human genome.Nat Rev Genet. 2012 Oct;13(10):678. doi: 10.1038/nrg3329. Epub 2012 Sep 7. Nat Rev Genet. 2012. PMID: 22955793 No abstract available.
Similar articles
-
The accessible chromatin landscape of the human genome.Nature. 2012 Sep 6;489(7414):75-82. doi: 10.1038/nature11232. Nature. 2012. PMID: 22955617 Free PMC article.
-
Global reference mapping of human transcription factor footprints.Nature. 2020 Jul;583(7818):729-736. doi: 10.1038/s41586-020-2528-x. Epub 2020 Jul 29. Nature. 2020. PMID: 32728250 Free PMC article.
-
An integrated encyclopedia of DNA elements in the human genome.Nature. 2012 Sep 6;489(7414):57-74. doi: 10.1038/nature11247. Nature. 2012. PMID: 22955616 Free PMC article.
-
Genomic footprinting.Nat Methods. 2016 Mar;13(3):213-21. doi: 10.1038/nmeth.3768. Nat Methods. 2016. PMID: 26914205 Review.
-
Genome-wide quantification of transcription factor binding at single-DNA-molecule resolution using methyl-transferase footprinting.Nat Protoc. 2021 Dec;16(12):5673-5706. doi: 10.1038/s41596-021-00630-1. Epub 2021 Nov 12. Nat Protoc. 2021. PMID: 34773120 Free PMC article. Review.
Cited by
-
Logical design of synthetic cis-regulatory DNA for genetic tracing of cell identities and state changes.Nat Commun. 2024 Feb 5;15(1):897. doi: 10.1038/s41467-024-45069-6. Nat Commun. 2024. PMID: 38316783 Free PMC article.
-
Time-Based Systems Biology Approaches to Capture and Model Dynamic Gene Regulatory Networks.Annu Rev Plant Biol. 2021 Jun 17;72:105-131. doi: 10.1146/annurev-arplant-081320-090914. Epub 2021 Mar 5. Annu Rev Plant Biol. 2021. PMID: 33667112 Free PMC article. Review.
-
Deciphering cis-regulatory control in inflammatory cells.Philos Trans R Soc Lond B Biol Sci. 2013 May 6;368(1620):20120370. doi: 10.1098/rstb.2012.0370. Print 2013. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 23650641 Free PMC article. Review.
-
Using the ENCODE Resource for Functional Annotation of Genetic Variants.Cold Spring Harb Protoc. 2015 Mar 11;2015(6):522-36. doi: 10.1101/pdb.top084988. Cold Spring Harb Protoc. 2015. PMID: 25762420 Free PMC article. Review.
-
A method for calculating probabilities of fitness consequences for point mutations across the human genome.Nat Genet. 2015 Mar;47(3):276-83. doi: 10.1038/ng.3196. Epub 2015 Jan 19. Nat Genet. 2015. PMID: 25599402 Free PMC article.
References
-
- Dynan WS, Tjian R. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell. 1983;35:79–87. - PubMed
-
- Gross DS, Garrard WT. Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 1988;57:159–197. - PubMed
-
- Thanos D, Maniatis T. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

