Analysis of T-DNA alleles of flavonoid biosynthesis genes in Arabidopsis ecotype Columbia
- PMID: 22947320
- PMCID: PMC3526476
- DOI: 10.1186/1756-0500-5-485
Analysis of T-DNA alleles of flavonoid biosynthesis genes in Arabidopsis ecotype Columbia
Abstract
Background: The flavonoid pathway is a long-standing and important tool for plant genetics, biochemistry, and molecular biology. Numerous flavonoid mutants have been identified in Arabidopsis over the past several decades in a variety of ecotypes. Here we present an analysis of Arabidopsis lines of ecotype Columbia carrying T-DNA insertions in genes encoding enzymes of the central flavonoid pathway. We also provide a comprehensive summary of various mutant alleles for these structural genes that have been described in the literature to date in a wide variety of ecotypes.
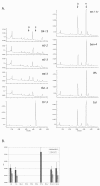
Findings: The confirmed knockout lines present easily-scorable phenotypes due to altered pigmentation of the seed coat (or testa). Knockouts for seven alleles for six flavonoid biosynthetic genes were confirmed by PCR and characterized by UPLC for altered flavonol content.
Conclusion: Seven mutant lines for six genes of the central flavonoid pathway were characterized in ecotype, Columbia. These lines represent a useful resource for integrating biochemical and physiological studies with genomic, transcriptomic, and proteomic data, much of which has been, and continues to be, generated in the Columbia background.
Figures



Similar articles
-
Disruption of specific flavonoid genes enhances the accumulation of flavonoid enzymes and end-products in Arabidopsis seedlings.Plant Mol Biol. 1999 May;40(1):45-54. doi: 10.1023/a:1026414301100. Plant Mol Biol. 1999. PMID: 10394944
-
Update on transparent testa mutants from Arabidopsis thaliana: characterisation of new alleles from an isogenic collection.Planta. 2014 Nov;240(5):955-70. doi: 10.1007/s00425-014-2088-0. Epub 2014 Jun 6. Planta. 2014. PMID: 24903359
-
A new dominant Arabidopsis transparent testa mutant, sk21-D, and modulation of seed flavonoid biosynthesis by KAN4.Plant Biotechnol J. 2010 Dec;8(9):979-93. doi: 10.1111/j.1467-7652.2010.00525.x. Plant Biotechnol J. 2010. PMID: 20444210
-
Increasing antioxidant levels in tomatoes through modification of the flavonoid biosynthetic pathway.J Exp Bot. 2002 Oct;53(377):2099-106. doi: 10.1093/jxb/erf044. J Exp Bot. 2002. PMID: 12324533 Review.
-
The flavonoid biosynthetic pathway in Arabidopsis: structural and genetic diversity.Plant Physiol Biochem. 2013 Nov;72:21-34. doi: 10.1016/j.plaphy.2013.02.001. Epub 2013 Feb 16. Plant Physiol Biochem. 2013. PMID: 23473981 Review.
Cited by
-
Genome-wide identification of grape ANS gene family and expression analysis at different fruit coloration stages.BMC Plant Biol. 2023 Dec 9;23(1):632. doi: 10.1186/s12870-023-04648-3. BMC Plant Biol. 2023. PMID: 38066449 Free PMC article.
-
PtoWRKY40 interacts with PtoPHR1-LIKE3 while regulating the phosphate starvation response in poplar.Plant Physiol. 2022 Nov 28;190(4):2688-2705. doi: 10.1093/plphys/kiac404. Plant Physiol. 2022. PMID: 36040189 Free PMC article.
-
A fast and simple LC-MS-based characterization of the flavonoid biosynthesis pathway for few seed(ling)s.BMC Plant Biol. 2016 Sep 1;16(1):190. doi: 10.1186/s12870-016-0880-7. BMC Plant Biol. 2016. PMID: 27586417 Free PMC article.
-
Isolating an active and inactive CACTA transposon from lettuce color mutants and characterizing their family.Plant Physiol. 2021 Jun 11;186(2):929-944. doi: 10.1093/plphys/kiab143. Plant Physiol. 2021. PMID: 33768232 Free PMC article.
-
Identification of MOS9 as an interaction partner for chalcone synthase in the nucleus.PeerJ. 2018 Sep 19;6:e5598. doi: 10.7717/peerj.5598. eCollection 2018. PeerJ. 2018. PMID: 30258711 Free PMC article.
References
-
- Winkel BSJ. In: The Science of Flavonoids. Grotewold E, editor. New York: Springer Science & Business Media; 2006. The biosynthesis of flavonoids; pp. 71–95.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

