JEnsembl: a version-aware Java API to Ensembl data systems
- PMID: 22945789
- PMCID: PMC3476335
- DOI: 10.1093/bioinformatics/bts525
JEnsembl: a version-aware Java API to Ensembl data systems
Abstract
Motivation: The Ensembl Project provides release-specific Perl APIs for efficient high-level programmatic access to data stored in various Ensembl database schema. Although Perl scripts are perfectly suited for processing large volumes of text-based data, Perl is not ideal for developing large-scale software applications nor embedding in graphical interfaces. The provision of a novel Java API would facilitate type-safe, modular, object-orientated development of new Bioinformatics tools with which to access, analyse and visualize Ensembl data.
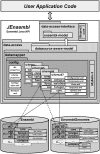
Results: The JEnsembl API implementation provides basic data retrieval and manipulation functionality from the Core, Compara and Variation databases for all species in Ensembl and EnsemblGenomes and is a platform for the development of a richer API to Ensembl datasources. The JEnsembl architecture uses a text-based configuration module to provide evolving, versioned mappings from database schema to code objects. A single installation of the JEnsembl API can therefore simultaneously and transparently connect to current and previous database instances (such as those in the public archive) thus facilitating better analysis repeatability and allowing 'through time' comparative analyses to be performed.
Availability: Project development, released code libraries, Maven repository and documentation are hosted at SourceForge (http://jensembl.sourceforge.net).
Figures




Similar articles
-
ArkMAP: integrating genomic maps across species and data sources.BMC Bioinformatics. 2013 Aug 13;14:246. doi: 10.1186/1471-2105-14-246. BMC Bioinformatics. 2013. PMID: 23941167 Free PMC article.
-
Ensembl Genomes: extending Ensembl across the taxonomic space.Nucleic Acids Res. 2010 Jan;38(Database issue):D563-9. doi: 10.1093/nar/gkp871. Epub 2009 Nov 1. Nucleic Acids Res. 2010. PMID: 19884133 Free PMC article.
-
The Ensembl REST API: Ensembl Data for Any Language.Bioinformatics. 2015 Jan 1;31(1):143-5. doi: 10.1093/bioinformatics/btu613. Epub 2014 Sep 17. Bioinformatics. 2015. PMID: 25236461 Free PMC article.
-
Interoperability with Moby 1.0--it's better than sharing your toothbrush!Brief Bioinform. 2008 May;9(3):220-31. doi: 10.1093/bib/bbn003. Epub 2008 Jan 31. Brief Bioinform. 2008. PMID: 18238804 Review.
-
The Ensembl core software libraries.Genome Res. 2004 May;14(5):929-33. doi: 10.1101/gr.1857204. Genome Res. 2004. PMID: 15123588 Free PMC article. Review.
Cited by
-
ArkMAP: integrating genomic maps across species and data sources.BMC Bioinformatics. 2013 Aug 13;14:246. doi: 10.1186/1471-2105-14-246. BMC Bioinformatics. 2013. PMID: 23941167 Free PMC article.
-
GMOL: An Interactive Tool for 3D Genome Structure Visualization.Sci Rep. 2016 Feb 12;6:20802. doi: 10.1038/srep20802. Sci Rep. 2016. PMID: 26868282 Free PMC article.

