DNA repair endonuclease ERCC1-XPF as a novel therapeutic target to overcome chemoresistance in cancer therapy
- PMID: 22941649
- PMCID: PMC3488251
- DOI: 10.1093/nar/gks818
DNA repair endonuclease ERCC1-XPF as a novel therapeutic target to overcome chemoresistance in cancer therapy
Abstract
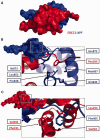
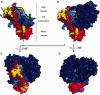
The ERCC1-XPF complex is a structure-specific endonuclease essential for the repair of DNA damage by the nucleotide excision repair pathway. It is also involved in other key cellular processes, including DNA interstrand crosslink (ICL) repair and DNA double-strand break (DSB) repair. New evidence has recently emerged, increasing our understanding of its requirement in these additional roles. In this review, we focus on the protein-protein and protein-DNA interactions made by the ERCC1 and XPF proteins and discuss how these coordinate ERCC1-XPF in its various roles. In a number of different cancers, high expression of ERCC1 has been linked to a poor response to platinum-based chemotherapy. We discuss prospects for the development of DNA repair inhibitors that target the activity, stability or protein interactions of the ERCC1-XPF complex as a novel therapeutic strategy to overcome chemoresistance.
Figures


Similar articles
-
Inhibition of the ERCC1-XPF structure-specific endonuclease to overcome cancer chemoresistance.DNA Repair (Amst). 2015 Jul;31:19-28. doi: 10.1016/j.dnarep.2015.04.002. Epub 2015 Apr 22. DNA Repair (Amst). 2015. PMID: 25956741
-
Multiple roles of the ERCC1-XPF endonuclease in DNA repair and resistance to anticancer drugs.Anticancer Res. 2010 Sep;30(9):3223-32. Anticancer Res. 2010. PMID: 20944091 Review.
-
Function and Interactions of ERCC1-XPF in DNA Damage Response.Molecules. 2018 Dec 5;23(12):3205. doi: 10.3390/molecules23123205. Molecules. 2018. PMID: 30563071 Free PMC article. Review.
-
The ERCC1 and ERCC4 (XPF) genes and gene products.Gene. 2015 Sep 15;569(2):153-61. doi: 10.1016/j.gene.2015.06.026. Epub 2015 Jun 12. Gene. 2015. PMID: 26074087 Free PMC article. Review.
-
Downregulation of XPF-ERCC1 enhances cisplatin efficacy in cancer cells.DNA Repair (Amst). 2010 Jul 1;9(7):745-53. doi: 10.1016/j.dnarep.2010.03.010. Epub 2010 Apr 24. DNA Repair (Amst). 2010. PMID: 20418188 Free PMC article.
Cited by
-
Demethoxycurcumin-Loaded Chitosan Nanoparticle Downregulates DNA Repair Pathway to Improve Cisplatin-Induced Apoptosis in Non-Small Cell Lung Cancer.Molecules. 2018 Dec 5;23(12):3217. doi: 10.3390/molecules23123217. Molecules. 2018. PMID: 30563166 Free PMC article.
-
DNA repair biomarkers XPF and phospho-MAPKAP kinase 2 correlate with clinical outcome in advanced head and neck cancer.PLoS One. 2014 Jul 14;9(7):e102112. doi: 10.1371/journal.pone.0102112. eCollection 2014. PLoS One. 2014. PMID: 25019640 Free PMC article.
-
Highlights of the DNA cutters: a short history of the restriction enzymes.Nucleic Acids Res. 2014 Jan;42(1):3-19. doi: 10.1093/nar/gkt990. Epub 2013 Oct 18. Nucleic Acids Res. 2014. PMID: 24141096 Free PMC article.
-
ATR pathway inhibition is synthetically lethal in cancer cells with ERCC1 deficiency.Cancer Res. 2014 May 15;74(10):2835-45. doi: 10.1158/0008-5472.CAN-13-3229. Epub 2014 Mar 24. Cancer Res. 2014. PMID: 24662920 Free PMC article.
-
Role of deubiquitinases in DNA damage response.DNA Repair (Amst). 2019 Apr;76:89-98. doi: 10.1016/j.dnarep.2019.02.011. Epub 2019 Feb 21. DNA Repair (Amst). 2019. PMID: 30831436 Free PMC article. Review.
References
-
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2nd edn. Washington, D.C., U.S.A: ASM Press; 2006.
-
- Aboussekhra A, Biggerstaff M, Shivji MK, Vilpo JA, Moncollin V, Podust VN, Protić M, Hübscher U, Egly JM, Wood RD. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 1995;80:859–868. - PubMed
-
- Naegeli H, Sugasawa K. The xeroderma pigmentosum pathway: decision tree analysis of DNA quality. DNA Repair. 2011;10:673–683. - PubMed
-
- Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 2008;9:958–970. - PubMed
-
- Camenisch U, Dip R, Schumacher SB, Schuler B, Naegeli H. Recognition of helical kinks by xeroderma pigmentosum group A protein triggers DNA excision repair. Nat. Struct. Mol. Biol. 2006;13:278–284. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

