S-bacillithiolation protects conserved and essential proteins against hypochlorite stress in firmicutes bacteria
- PMID: 22938038
- PMCID: PMC3584511
- DOI: 10.1089/ars.2012.4686
S-bacillithiolation protects conserved and essential proteins against hypochlorite stress in firmicutes bacteria
Abstract
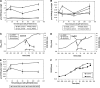
Aims: Protein S-bacillithiolations are mixed disulfides between protein thiols and the bacillithiol (BSH) redox buffer that occur in response to NaOCl in Bacillus subtilis. We used BSH-specific immunoblots, shotgun liquid chromatography (LC)-tandem mass spectrometry (MS/MS) analysis and redox proteomics to characterize the S-bacillithiolomes of B. subtilis, B. megaterium, B. pumilus, B. amyloliquefaciens, and Staphylococcus carnosus and also measured the BSH/oxidized bacillithiol disulfide (BSSB) redox ratio after NaOCl stress.
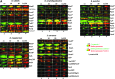
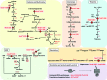
Results: In total, 54 proteins with characteristic S-bacillithiolation (SSB) sites were identified, including 29 unique proteins and eight proteins conserved in two or more of these bacteria. The methionine synthase MetE is the most abundant S-bacillithiolated protein in Bacillus species after NaOCl exposure. Further, S-bacillithiolated proteins include the translation elongation factor EF-Tu and aminoacyl-tRNA synthetases (ThrS), the DnaK and GrpE chaperones, the two-Cys peroxiredoxin YkuU, the ferredoxin-NADP(+) oxidoreductase YumC, the inorganic pyrophosphatase PpaC, the inosine-5'-monophosphate dehydrogenase GuaB, proteins involved in thiamine biosynthesis (ThiG and ThiM), queuosine biosynthesis (QueF), biosynthesis of aromatic amino acids (AroA and AroE), serine (SerA), branched-chain amino acids (YwaA), and homocysteine (LuxS and MetI). The thioredoxin-like proteins, YphP and YtxJ, are S-bacillithiolated at their active sites, suggesting a function in the de-bacillithiolation process. S-bacillithiolation is accompanied by a two-fold increase in the BSSB level and a decrease in the BSH/BSSB redox ratio in B. subtilis.
Innovation: Many essential and conserved proteins, including the dominant MetE, were identified in the S-bacillithiolome of different Bacillus species and S. carnosus using shotgun-LC-MS/MS analyses.
Conclusion: S-bacillithiolation is a widespread redox control mechanism among Firmicutes bacteria that protects conserved metabolic enzymes and essential proteins against overoxidation.
Figures



Similar articles
-
S-bacillithiolation protects against hypochlorite stress in Bacillus subtilis as revealed by transcriptomics and redox proteomics.Mol Cell Proteomics. 2011 Nov;10(11):M111.009506. doi: 10.1074/mcp.M111.009506. Epub 2011 Jul 11. Mol Cell Proteomics. 2011. PMID: 21749987 Free PMC article.
-
Redox regulation in Bacillus subtilis: The bacilliredoxins BrxA(YphP) and BrxB(YqiW) function in de-bacillithiolation of S-bacillithiolated OhrR and MetE.Antioxid Redox Signal. 2014 Jul 20;21(3):357-67. doi: 10.1089/ars.2013.5327. Epub 2014 Mar 13. Antioxid Redox Signal. 2014. PMID: 24313874 Free PMC article.
-
Protein S-Bacillithiolation Functions in Thiol Protection and Redox Regulation of the Glyceraldehyde-3-Phosphate Dehydrogenase Gap in Staphylococcus aureus Under Hypochlorite Stress.Antioxid Redox Signal. 2018 Feb 20;28(6):410-430. doi: 10.1089/ars.2016.6897. Epub 2017 Jan 18. Antioxid Redox Signal. 2018. PMID: 27967218 Free PMC article.
-
The Role of Bacillithiol in Gram-Positive Firmicutes.Antioxid Redox Signal. 2018 Feb 20;28(6):445-462. doi: 10.1089/ars.2017.7057. Epub 2017 Apr 24. Antioxid Redox Signal. 2018. PMID: 28301954 Free PMC article. Review.
-
Redox regulation by reversible protein S-thiolation in bacteria.Front Microbiol. 2015 Mar 16;6:187. doi: 10.3389/fmicb.2015.00187. eCollection 2015. Front Microbiol. 2015. PMID: 25852656 Free PMC article. Review.
Cited by
-
Importance of bacillithiol in the oxidative stress response of Staphylococcus aureus.Infect Immun. 2014 Jan;82(1):316-32. doi: 10.1128/IAI.01074-13. Epub 2013 Oct 28. Infect Immun. 2014. PMID: 24166956 Free PMC article.
-
Redox-Sensing Under Hypochlorite Stress and Infection Conditions by the Rrf2-Family Repressor HypR in Staphylococcus aureus.Antioxid Redox Signal. 2018 Sep 1;29(7):615-636. doi: 10.1089/ars.2017.7354. Epub 2018 Jan 30. Antioxid Redox Signal. 2018. PMID: 29237286 Free PMC article.
-
Glutathionylspermidine in the modification of protein SH groups: the enzymology and its application to study protein glutathionylation.Molecules. 2015 Jan 15;20(1):1452-74. doi: 10.3390/molecules20011452. Molecules. 2015. PMID: 25599150 Free PMC article. Review.
-
The Disulfide Stress Response and Protein S-thioallylation Caused by Allicin and Diallyl Polysulfanes in Bacillus subtilis as Revealed by Transcriptomics and Proteomics.Antioxidants (Basel). 2019 Nov 29;8(12):605. doi: 10.3390/antiox8120605. Antioxidants (Basel). 2019. PMID: 31795512 Free PMC article.
-
The Bacillus subtilis monothiol bacilliredoxin BrxC (YtxJ) and the Bdr (YpdA) disulfide reductase reduce S-bacillithiolated proteins.Redox Biol. 2021 Jun;42:101935. doi: 10.1016/j.redox.2021.101935. Epub 2021 Mar 6. Redox Biol. 2021. PMID: 33722570 Free PMC article.
References
-
- Caldas TD. El Yaagoubi A. Richarme G. Chaperone properties of bacterial elongation factor EF-Tu. J Biol Chem. 1998;273:11478–11482. - PubMed
-
- Chen XH. Koumoutsi A. Scholz R. Eisenreich A. Schneider K. Heinemeyer I. Morgenstern B. Voss B. Hess WR. Reva O. Junge H. Voigt B. Jungblut PR. Vater J. Sussmuth R. Liesegang H. Strittmatter A. Gottschalk G. Borriss R. Comparative analysis of the complete genome sequence of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42. Nat Biotechnol. 2007;25:1007–1014. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

