A hypermorphic SP1-binding CD24 variant associates with risk and progression of multiple sclerosis
- PMID: 22937211
- PMCID: PMC3426393
A hypermorphic SP1-binding CD24 variant associates with risk and progression of multiple sclerosis
Abstract
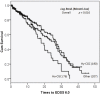
A large number of risk alleles have been identified for multiple sclerosis (MS). However, how genetic variations may affect pathogenesis remains largely unknown for most risk alleles. Through direct sequencing of CD24 promoter region, we identified a cluster of 7 new single nucleotide polymorphisms in the CD24 promoter. A hypermorphic haplotype consisting of 3 SNPs was identified through association studies consisting of 935 control and 764 MS patients (P=0.001, odds ratio 1.3). The variant is also associated with more rapid progression of MS (P=0.016, log rank test). In cells that are heterozygous for the risk allele, chromatin immunoprecipitation revealed that risk allele specifically bind to a transcription factor SP1, which is selectively required for the hypermorphic promoter activity of the variant. In MS patients, the CD24 transcript levels associate with the SP1-binding variant in a dose-dependent manner (P=7x10(-4)). Our data revealed a potential role for SP1-mediated transcriptional regulation in MS pathogenesis.
Keywords: Multiple sclerosis (MS); SP-binding CD24; promoter; risk alleles; single nucleotide polymorphisms (SNP).
Figures



Similar articles
-
Association between CD24-P226-C/T polymorphism and multiple sclerosis: a meta-analysis.Immunol Invest. 2015;44(4):321-30. doi: 10.3109/08820139.2014.1003650. Immunol Invest. 2015. PMID: 25942344
-
A dinucleotide deletion in CD24 confers protection against autoimmune diseases.PLoS Genet. 2007 Apr 6;3(4):e49. doi: 10.1371/journal.pgen.0030049. Epub 2007 Feb 21. PLoS Genet. 2007. PMID: 17411341 Free PMC article.
-
CD24 is a genetic modifier for risk and progression of multiple sclerosis.Proc Natl Acad Sci U S A. 2003 Dec 9;100(25):15041-6. doi: 10.1073/pnas.2533866100. Epub 2003 Dec 1. Proc Natl Acad Sci U S A. 2003. PMID: 14657362 Free PMC article.
-
The -318 C>G single-nucleotide polymorphism in GNAI2 gene promoter region impairs transcriptional activity through specific binding of Sp1 transcription factor and is associated with high blood pressure in Caucasians from Italy.J Am Soc Nephrol. 2006 Apr;17(4 Suppl 2):S115-9. doi: 10.1681/ASN.2005121340. J Am Soc Nephrol. 2006. PMID: 16565233
-
Polymorphisms of the CD24 Gene Are Associated with Risk of Multiple Sclerosis: A Meta-Analysis.Int J Mol Sci. 2015 Jun 1;16(6):12368-81. doi: 10.3390/ijms160612368. Int J Mol Sci. 2015. PMID: 26039238 Free PMC article.
Cited by
-
Curcumin Suppresses Metastasis via Sp-1, FAK Inhibition, and E-Cadherin Upregulation in Colorectal Cancer.Evid Based Complement Alternat Med. 2013;2013:541695. doi: 10.1155/2013/541695. Epub 2013 Jul 21. Evid Based Complement Alternat Med. 2013. PMID: 23970932 Free PMC article.
-
Nucleophosmin/B23 promotes endometrial cancer cell escape from macrophage phagocytosis by increasing CD24 expression.J Mol Med (Berl). 2021 Aug;99(8):1125-1137. doi: 10.1007/s00109-021-02079-x. Epub 2021 May 5. J Mol Med (Berl). 2021. PMID: 33954835
-
CD24 is a genetic modifier for risk and progression of prostate cancer.Mol Carcinog. 2017 Feb;56(2):641-650. doi: 10.1002/mc.22522. Epub 2016 Jul 19. Mol Carcinog. 2017. PMID: 27377469 Free PMC article.
-
A CD24-p53 axis contributes to African American prostate cancer disparities.Prostate. 2020 May;80(8):609-618. doi: 10.1002/pros.23973. Epub 2020 Mar 13. Prostate. 2020. PMID: 32168400 Free PMC article.
-
Detecting rare haplotype-environment interaction with logistic Bayesian LASSO.Genet Epidemiol. 2014 Jan;38(1):31-41. doi: 10.1002/gepi.21773. Epub 2013 Nov 23. Genet Epidemiol. 2014. PMID: 24272913 Free PMC article.
References
-
- Ebers GC, Sadovnick AD, Risch NJ. A genetic basis for familial aggregation in multiple sclerosis. Canadian Collaborative Study Group. Nature. 1995;377:150–151. - PubMed
-
- Ramagopalan SV, Ebers GC. Genes for multiple sclerosis. Lancet. 2008;371:283–285. - PubMed
-
- Bahlo M, Booth DR, Broadley SA, Brown MA, Foote SJ, Griffiths LR, Kilpatrick TJ, Lechner-Scott J, Moscato P, Perreau VM, Rubio JP, Scott RJ, Stankovich J, Stewart GJ, Taylor BV, Wiley J, Brown MA, Booth DR, Clarke G, Cox MB, Csurhes PA, Danoy P, Drysdale K, Field J, Foote SJ, Greer JM, Griffiths LR, Guru P, Hadler J, McMorran BJ, Jensen CJ, Johnson LJ, McCallum R, Merriman M, Merriman T, Pryce K, Scott RJ, Stewart GJ, Tajouri L, Wilkins EJ, Rubio JP, Bahlo M, Brown MA, Browning BL, Browning SR, Perera D, Rubio JP, Stankovich J, Broadley S, Butzkueven H, Carroll WM, Chapman C, Kermode AG, Marriott M, Mason D, Heard RN, Pender MP, Slee M, Tubridy N, Lechner-Scott J, Taylor BV, Willoughby E, Kilpatrick TJ. Genome-wide association study identifies new multiple sclerosis susceptibility loci on chromosomes 12 and 20. Nat Genet. 2009;41:824–828. - PubMed
-
- Aulchenko YS, Hoppenbrouwers IA, Ramagopalan SV, Broer L, Jafari N, Hillert J, Link J, Lundstrom W, Greiner E, Dessa Sadovnick A, Goossens D, Van Broeckhoven C, Del-Favero J, Ebers GC, Oostra BA, van Duijn CM, Hintzen RQ. Genetic variation in the KIF1B locus influences susceptibility to multiple sclerosis. Nat Genet. 2008;40:1402–1403. - PubMed
-
- Baranzini SE, Wang J, Gibson RA, Galwey N, Naegelin Y, Barkhof F, Radue EW, Lindberg RL, Uitdehaag BM, Johnson MR, Angelakopoulou A, Hall L, Richardson JC, Prinjha RK, Gass A, Geurts JJ, Kragt J, Sombekke M, Vrenken H, Qualley P, Lincoln RR, Gomez R, Caillier SJ, George MF, Mousavi H, Guerrero R, Okuda DT, Cree BA, Green AJ, Waubant E, Goodin DS, Pelletier D, Matthews PM, Hauser SL, Kappos L, Polman CH, Oksenberg JR. Genome-wide association analysis of susceptibility and clinical phenotype in multiple sclerosis. Hum Mol Genet. 2009;18:767–778. - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
