Interferon regulatory factor 6 promotes differentiation of the periderm by activating expression of Grainyhead-like 3
- PMID: 22931925
- PMCID: PMC3541433
- DOI: 10.1038/jid.2012.269
Interferon regulatory factor 6 promotes differentiation of the periderm by activating expression of Grainyhead-like 3
Erratum in
- J Invest Dermatol. 2013 Mar;133(3):859
Abstract
IFN regulatory factor 6 (IRF6) is a transcription factor that, in mammals, is required for the differentiation of skin, breast epithelium, and oral epithelium. However, the transcriptional targets that mediate these effects are currently unknown. In zebrafish and frog embryos, Irf6 is necessary for differentiation of the embryonic superficial epithelium, or periderm. Here we use microarrays to identify genes that are expressed in the zebrafish periderm and whose expression is inhibited by a dominant-negative variant of Irf6 (dnIrf6). These methods identify Grainyhead-like 3 (Grhl3), an ancient regulator of the epidermal permeability barrier, as acting downstream of Irf6. In human keratinocytes, IRF6 binds conserved elements near the GRHL3 [corrected] promoter. We show that one of these elements has enhancer activity in human keratinocytes and zebrafish periderm, suggesting that Irf6 directly stimulates Grhl3 expression in these tissues. Simultaneous inhibition of grhl1 and grhl3 disrupts periderm differentiation in zebrafish, and, intriguingly, forced grhl3 expression restores periderm markers in both zebrafish injected with dnIrf6 and frog embryos depleted of Irf6. Finally, in Irf6-deficient mouse embryos, Grhl3 expression in the periderm and oral epithelium is virtually absent. These results indicate that Grhl3 is a key effector of Irf6 in periderm differentiation.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
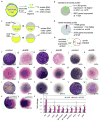
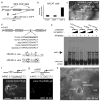
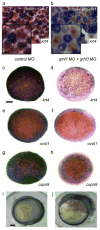
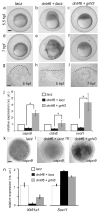
Similar articles
-
Dominant mutations in GRHL3 cause Van der Woude Syndrome and disrupt oral periderm development.Am J Hum Genet. 2014 Jan 2;94(1):23-32. doi: 10.1016/j.ajhg.2013.11.009. Epub 2013 Dec 19. Am J Hum Genet. 2014. PMID: 24360809 Free PMC article.
-
Irf6 directly regulates Klf17 in zebrafish periderm and Klf4 in murine oral epithelium, and dominant-negative KLF4 variants are present in patients with cleft lip and palate.Hum Mol Genet. 2016 Feb 15;25(4):766-76. doi: 10.1093/hmg/ddv614. Epub 2015 Dec 21. Hum Mol Genet. 2016. PMID: 26692521 Free PMC article.
-
Maternal Interferon Regulatory Factor 6 is required for the differentiation of primary superficial epithelia in Danio and Xenopus embryos.Dev Biol. 2009 Jan 1;325(1):249-62. doi: 10.1016/j.ydbio.2008.10.031. Epub 2008 Nov 5. Dev Biol. 2009. PMID: 19013452 Free PMC article.
-
Toward an orofacial gene regulatory network.Dev Dyn. 2016 Mar;245(3):220-32. doi: 10.1002/dvdy.24341. Epub 2015 Sep 17. Dev Dyn. 2016. PMID: 26332872 Free PMC article. Review.
-
Periderm: Life-cycle and function during orofacial and epidermal development.Semin Cell Dev Biol. 2019 Jul;91:75-83. doi: 10.1016/j.semcdb.2017.08.021. Epub 2017 Aug 10. Semin Cell Dev Biol. 2019. PMID: 28803895 Review.
Cited by
-
A Novel Combined Scientific and Artistic Approach for the Advanced Characterization of Interactomes: The Akirin/Subolesin Model.Vaccines (Basel). 2020 Feb 8;8(1):77. doi: 10.3390/vaccines8010077. Vaccines (Basel). 2020. PMID: 32046307 Free PMC article.
-
The RIPK4-IRF6 signalling axis safeguards epidermal differentiation and barrier function.Nature. 2019 Oct;574(7777):249-253. doi: 10.1038/s41586-019-1615-3. Epub 2019 Oct 2. Nature. 2019. PMID: 31578523
-
Grainyhead-like (Grhl) Target Genes in Development and Cancer.Int J Mol Sci. 2022 Mar 1;23(5):2735. doi: 10.3390/ijms23052735. Int J Mol Sci. 2022. PMID: 35269877 Free PMC article. Review.
-
Understanding the development of oral epithelial organs through single cell transcriptomic analysis.Development. 2022 Aug 15;149(16):dev200539. doi: 10.1242/dev.200539. Epub 2022 Aug 17. Development. 2022. PMID: 35831953 Free PMC article.
-
Dominant mutations in GRHL3 cause Van der Woude Syndrome and disrupt oral periderm development.Am J Hum Genet. 2014 Jan 2;94(1):23-32. doi: 10.1016/j.ajhg.2013.11.009. Epub 2013 Dec 19. Am J Hum Genet. 2014. PMID: 24360809 Free PMC article.
References
-
- Auden A, Caddy J, Wilanowski T, et al. Spatial and temporal expression of the Grainyhead-like transcription factor family during murine development. Gene Expr Patterns. 2006;6(8):964–970. - PubMed
-
- Bakkers J, Hild M, Kramer C, et al. Zebrafish DeltaNp63 is a direct target of Bmp signaling and encodes a transcriptional repressor blocking neural specification in the ventral ectoderm. Dev Cell. 2002;2(5):617–627. - PubMed
-
- Boglev Y, Wilanowski T, Caddy J, et al. The unique and cooperative roles of the Grainy head-like transcription factors in epidermal development reflect unexpected target gene specificity. Developmental biology. 2011;349(2):512–522. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 DE008559/DE/NIDCR NIH HHS/United States
- R01GM77429/GM/NIGMS NIH HHS/United States
- F31 DE022696/DE/NIDCR NIH HHS/United States
- GM08399904/GM/NIGMS NIH HHS/United States
- R01 DE021071/DE/NIDCR NIH HHS/United States
- DE13513/DE/NIDCR NIH HHS/United States
- GM067841/GM/NIGMS NIH HHS/United States
- T32 GM008629/GM/NIGMS NIH HHS/United States
- R01 GM083999/GM/NIGMS NIH HHS/United States
- R01 GM067841/GM/NIGMS NIH HHS/United States
- 5T32DC000040-17/DC/NIDCD NIH HHS/United States
- R01 GM077429/GM/NIGMS NIH HHS/United States
- T32 DC000040/DC/NIDCD NIH HHS/United States
- R01 DE013513/DE/NIDCR NIH HHS/United States
- DE21071/DE/NIDCR NIH HHS/United States
- T32 GM082729/GM/NIGMS NIH HHS/United States
- 5R37DE008559-21/DE/NIDCR NIH HHS/United States
- 1F31DE022696-01/DE/NIDCR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

