LAB-1 targets PP1 and restricts Aurora B kinase upon entrance into meiosis to promote sister chromatid cohesion
- PMID: 22927794
- PMCID: PMC3424243
- DOI: 10.1371/journal.pbio.1001378
LAB-1 targets PP1 and restricts Aurora B kinase upon entrance into meiosis to promote sister chromatid cohesion
Abstract
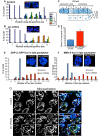
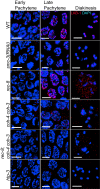
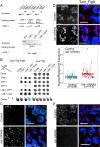
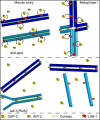
Successful execution of the meiotic program depends on the timely establishment and removal of sister chromatid cohesion. LAB-1 has been proposed to act in the latter by preventing the premature removal of the meiosis-specific cohesin REC-8 at metaphase I in C. elegans, yet the mechanism and scope of LAB-1 function remained unknown. Here we identify an unexpected earlier role for LAB-1 in promoting the establishment of sister chromatid cohesion in prophase I. LAB-1 and REC-8 are both required for the chromosomal association of the cohesin complex subunit SMC-3. Depletion of lab-1 results in partial loss of sister chromatid cohesion in rec-8 and coh-4 coh-3 mutants and further enhanced chromatid dissociation in worms where all three kleisins are mutated. Moreover, lab-1 depletion results in increased Aurora B kinase (AIR-2) signals in early prophase I nuclei, coupled with a parallel decrease in signals for the PP1 homolog, GSP-2. Finally, LAB-1 directly interacts with GSP-1 and GSP-2. We propose that LAB-1 targets the PP1 homologs to the chromatin at the onset of meiosis I, thereby antagonizing AIR-2 and cooperating with the cohesin complex to promote sister chromatid association and normal progression of the meiotic program.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
LAB-1 antagonizes the Aurora B kinase in C. elegans.Genes Dev. 2008 Oct 15;22(20):2869-85. doi: 10.1101/gad.1691208. Genes Dev. 2008. PMID: 18923084 Free PMC article.
-
A Novel Role for α-Importins and Akirin in Establishment of Meiotic Sister Chromatid Cohesion in Caenorhabditis elegans.Genetics. 2019 Feb;211(2):617-635. doi: 10.1534/genetics.118.301458. Epub 2018 Dec 18. Genetics. 2019. PMID: 30563860 Free PMC article.
-
Spatiotemporal regulation of Aurora B recruitment ensures release of cohesion during C. elegans oocyte meiosis.Nat Commun. 2018 Feb 26;9(1):834. doi: 10.1038/s41467-018-03229-5. Nat Commun. 2018. PMID: 29483514 Free PMC article.
-
Cohesin complexes and sister chromatid cohesion in mammalian meiosis.Genome Dyn. 2009;5:94-116. doi: 10.1159/000166622. Genome Dyn. 2009. PMID: 18948710 Review.
-
The expanding phenotypes of cohesinopathies: one ring to rule them all!Cell Cycle. 2019 Nov;18(21):2828-2848. doi: 10.1080/15384101.2019.1658476. Epub 2019 Sep 13. Cell Cycle. 2019. PMID: 31516082 Free PMC article. Review.
Cited by
-
Condensin I protects meiotic cohesin from WAPL-1 mediated removal.PLoS Genet. 2018 May 16;14(5):e1007382. doi: 10.1371/journal.pgen.1007382. eCollection 2018 May. PLoS Genet. 2018. PMID: 29768402 Free PMC article.
-
ZTF-8 interacts with the 9-1-1 complex and is required for DNA damage response and double-strand break repair in the C. elegans germline.PLoS Genet. 2014 Oct 16;10(10):e1004723. doi: 10.1371/journal.pgen.1004723. eCollection 2014 Oct. PLoS Genet. 2014. PMID: 25329393 Free PMC article.
-
Analysis of meiosis in Pristionchus pacificus reveals plasticity in homolog pairing and synapsis in the nematode lineage.Elife. 2021 Aug 24;10:e70990. doi: 10.7554/eLife.70990. Elife. 2021. PMID: 34427184 Free PMC article.
-
GRAS-1 is a novel regulator of early meiotic chromosome dynamics in C. elegans.PLoS Genet. 2023 Feb 21;19(2):e1010666. doi: 10.1371/journal.pgen.1010666. eCollection 2023 Feb. PLoS Genet. 2023. PMID: 36809245 Free PMC article.
-
TOP-2 is differentially required for the proper maintenance of the cohesin subunit REC-8 on meiotic chromosomes in Caenorhabditis elegans spermatogenesis and oogenesis.Genetics. 2022 Sep 30;222(2):iyac120. doi: 10.1093/genetics/iyac120. Genetics. 2022. PMID: 35951744 Free PMC article.
References
-
- Hassold T, Hunt P (2001) To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet 2: 280–291. - PubMed
-
- Kops G, Weaver B, Cleveland D (2005) On the road to cancer: aneuploidy and the mitotic checkpoint. Nat Rev Cancer 5: 773–785. - PubMed
-
- Nasmyth K (2001) Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet 35: 673–745. - PubMed
-
- Uhlmann F (2003) Chromosome cohesion and separation: from men and molecules. Curr Biol 13: R104–R114. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

