Near-atomic resolution structural model of the yeast 26S proteasome
- PMID: 22927375
- PMCID: PMC3443124
- DOI: 10.1073/pnas.1213333109
Near-atomic resolution structural model of the yeast 26S proteasome
Abstract
The 26S proteasome operates at the executive end of the ubiquitin-proteasome pathway. Here, we present a cryo-EM structure of the Saccharomyces cerevisiae 26S proteasome at a resolution of 7.4 Å or 6.7 Å (Fourier-Shell Correlation of 0.5 or 0.3, respectively). We used this map in conjunction with molecular dynamics-based flexible fitting to build a near-atomic resolution model of the holocomplex. The quality of the map allowed us to assign α-helices, the predominant secondary structure element of the regulatory particle subunits, throughout the entire map. We were able to determine the architecture of the Rpn8/Rpn11 heterodimer, which had hitherto remained elusive. The MPN domain of Rpn11 is positioned directly above the AAA-ATPase N-ring suggesting that Rpn11 deubiquitylates substrates immediately following commitment and prior to their unfolding by the AAA-ATPase module. The MPN domain of Rpn11 dimerizes with that of Rpn8 and the C-termini of both subunits form long helices, which are integral parts of a coiled-coil module. Together with the C-terminal helices of the six PCI-domain subunits they form a very large coiled-coil bundle, which appears to serve as a flexible anchoring device for all the lid subunits.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



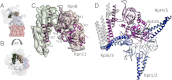
Similar articles
-
Base-CP proteasome can serve as a platform for stepwise lid formation.Biosci Rep. 2015 Jan 27;35(3):e00194. doi: 10.1042/BSR20140173. Biosci Rep. 2015. PMID: 26182356 Free PMC article.
-
Solution structure of yeast Rpn9: insights into proteasome lid assembly.J Biol Chem. 2015 Mar 13;290(11):6878-89. doi: 10.1074/jbc.M114.626762. Epub 2015 Jan 28. J Biol Chem. 2015. PMID: 25631053 Free PMC article.
-
Crystal structure of the proteasomal deubiquitylation module Rpn8-Rpn11.Proc Natl Acad Sci U S A. 2014 Feb 25;111(8):2984-9. doi: 10.1073/pnas.1400546111. Epub 2014 Feb 10. Proc Natl Acad Sci U S A. 2014. PMID: 24516147 Free PMC article.
-
Molecular and cellular dynamics of the 26S proteasome.Biochim Biophys Acta Proteins Proteom. 2021 Mar;1869(3):140583. doi: 10.1016/j.bbapap.2020.140583. Epub 2020 Dec 13. Biochim Biophys Acta Proteins Proteom. 2021. PMID: 33321258 Review.
-
Structure, Dynamics and Function of the 26S Proteasome.Subcell Biochem. 2021;96:1-151. doi: 10.1007/978-3-030-58971-4_1. Subcell Biochem. 2021. PMID: 33252727 Review.
Cited by
-
Structural characterization of the interaction of Ubp6 with the 26S proteasome.Proc Natl Acad Sci U S A. 2015 Jul 14;112(28):8626-31. doi: 10.1073/pnas.1510449112. Epub 2015 Jun 30. Proc Natl Acad Sci U S A. 2015. PMID: 26130806 Free PMC article.
-
An assay for 26S proteasome activity based on fluorescence anisotropy measurements of dye-labeled protein substrates.Anal Biochem. 2016 Sep 15;509:50-59. doi: 10.1016/j.ab.2016.05.026. Epub 2016 Jun 11. Anal Biochem. 2016. PMID: 27296635 Free PMC article.
-
The C-terminal residues of Saccharomyces cerevisiae Mec1 are required for its localization, stability, and function.G3 (Bethesda). 2013 Oct 3;3(10):1661-74. doi: 10.1534/g3.113.006841. G3 (Bethesda). 2013. PMID: 23934994 Free PMC article.
-
Structural and biochemical properties of an extreme 'salt-loving' proteasome activating nucleotidase from the archaeon Haloferax volcanii.Extremophiles. 2014 Mar;18(2):283-93. doi: 10.1007/s00792-013-0615-8. Epub 2013 Dec 17. Extremophiles. 2014. PMID: 24343376 Free PMC article.
-
The Not4 E3 ligase and CCR4 deadenylase play distinct roles in protein quality control.PLoS One. 2014 Jan 17;9(1):e86218. doi: 10.1371/journal.pone.0086218. eCollection 2014. PLoS One. 2014. PMID: 24465968 Free PMC article.
References
-
- Voges D, Zwickl P, Baumeister W. The 26S proteasome: A molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–1068. - PubMed
-
- Groll M, et al. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–471. - PubMed
-
- Lowe J, et al. Crystal structure of the 20S proteasome from the archaeon T.acidophilum at 3.4 A resolution. Science. 1995;268:533–539. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

