Genomic profiling of a human organotypic model of AEC syndrome reveals ZNF750 as an essential downstream target of mutant TP63
- PMID: 22922031
- PMCID: PMC3511987
- DOI: 10.1016/j.ajhg.2012.07.007
Genomic profiling of a human organotypic model of AEC syndrome reveals ZNF750 as an essential downstream target of mutant TP63
Abstract
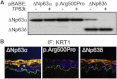
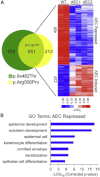
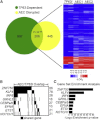
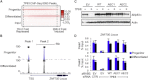
The basis for impaired differentiation in TP63 mutant ankyloblepharon-ectodermal dysplasia-clefting (AEC) syndrome is unknown. Human epidermis harboring AEC TP63 mutants recapitulated this impairment, along with downregulation of differentiation activators, including HOPX, GRHL3, KLF4, PRDM1, and ZNF750. Gene-set enrichment analysis indicated that disrupted expression of epidermal differentiation programs under the control of ZNF750 and KLF4 accounted for the majority of disrupted epidermal differentiation resulting from AEC mutant TP63. Chromatin immunoprecipitation (ChIP) analysis and ChIP-sequencing of TP63 binding in differentiated keratinocytes revealed ZNF750 as a direct target of wild-type and AEC mutant TP63. Restoring ZNF750 to AEC model tissue rescued activator expression and differentiation, indicating that AEC TP63-mediated ZNF750 inhibition contributes to differentiation defects in AEC. Incorporating disease-causing mutants into regenerated human tissue can thus dissect pathomechanisms and identify targets that reverse disease features.
Copyright © 2012 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Novel missense mutation of the TP63 gene in a newborn with Hay-Wells/Ankyloblepharon-Ectodermal defects-Cleft lip/palate (AEC) syndrome: clinical report and follow-up.Ital J Pediatr. 2021 Sep 28;47(1):196. doi: 10.1186/s13052-021-01152-y. Ital J Pediatr. 2021. PMID: 34583755 Free PMC article.
-
Novel variant in the TP63 gene associated to ankyloblepharon-ectodermal dysplasia-cleft lip/palate (AEC) syndrome.Ophthalmic Genet. 2017 May-Jun;38(3):277-280. doi: 10.1080/13816810.2016.1210649. Epub 2016 Aug 2. Ophthalmic Genet. 2017. PMID: 27485918
-
Tooth defects of EEC and AEC syndrome caused by heterozygous TP63 mutations in three Chinese families and genotype-phenotype correlation analyses of TP63-related disorders.Mol Genet Genomic Med. 2019 Jun;7(6):e704. doi: 10.1002/mgg3.704. Epub 2019 May 2. Mol Genet Genomic Med. 2019. PMID: 31050217 Free PMC article.
-
Modeling AEC-New approaches to study rare genetic disorders.Am J Med Genet A. 2014 Oct;164A(10):2443-54. doi: 10.1002/ajmg.a.36455. Epub 2014 Mar 24. Am J Med Genet A. 2014. PMID: 24665072 Free PMC article. Review.
-
TP63-related disorders: two case reports and a brief review of the literature.Dermatol Online J. 2021 Nov 15;27(11). doi: 10.5070/D3271156088. Dermatol Online J. 2021. PMID: 35130400 Review.
Cited by
-
Transcriptome analysis of psoriasis in a large case-control sample: RNA-seq provides insights into disease mechanisms.J Invest Dermatol. 2014 Jul;134(7):1828-1838. doi: 10.1038/jid.2014.28. Epub 2014 Jan 17. J Invest Dermatol. 2014. PMID: 24441097 Free PMC article.
-
A Human Stem Cell-Based System to Study the Role of TP63 Mutations in Ectodermal Dysplasias.J Invest Dermatol. 2018 Jul;138(7):1662-1665. doi: 10.1016/j.jid.2018.02.016. Epub 2018 Feb 23. J Invest Dermatol. 2018. PMID: 29481901 Free PMC article. No abstract available.
-
ΔNp63α Promotes Breast Cancer Cell Motility through the Selective Activation of Components of the Epithelial-to-Mesenchymal Transition Program.Cancer Res. 2015 Sep 15;75(18):3925-35. doi: 10.1158/0008-5472.CAN-14-3363. Epub 2015 Aug 19. Cancer Res. 2015. PMID: 26292362 Free PMC article.
-
p63 Transcription Factor Regulates Nuclear Shape and Expression of Nuclear Envelope-Associated Genes in Epidermal Keratinocytes.J Invest Dermatol. 2017 Oct;137(10):2157-2167. doi: 10.1016/j.jid.2017.05.013. Epub 2017 Jun 6. J Invest Dermatol. 2017. PMID: 28595999 Free PMC article.
-
Dissecting the DNA binding landscape and gene regulatory network of p63 and p53.Elife. 2020 Dec 2;9:e63266. doi: 10.7554/eLife.63266. Elife. 2020. PMID: 33263276 Free PMC article.
References
-
- Mills A.A., Zheng B., Wang X.J., Vogel H., Roop D.R., Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. - PubMed
-
- Yang A., Schweitzer R., Sun D., Kaghad M., Walker N., Bronson R.T., Tabin C., Sharpe A., Caput D., Crum C., McKeon F. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. - PubMed
-
- Senoo M., Pinto F., Crum C.P., McKeon F. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129:523–536. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

