Conservation of caspase substrates across metazoans suggests hierarchical importance of signaling pathways over specific targets and cleavage site motifs in apoptosis
- PMID: 22918439
- PMCID: PMC3504717
- DOI: 10.1038/cdd.2012.99
Conservation of caspase substrates across metazoans suggests hierarchical importance of signaling pathways over specific targets and cleavage site motifs in apoptosis
Abstract
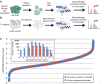
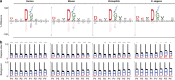
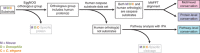
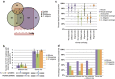
Caspases, cysteine proteases with aspartate specificity, are key players in programmed cell death across the metazoan lineage. Hundreds of apoptotic caspase substrates have been identified in human cells. Some have been extensively characterized, revealing key functional nodes for apoptosis signaling and important drug targets in cancer. But the functional significance of most cuts remains mysterious. We set out to better understand the importance of caspase cleavage specificity in apoptosis by asking which cleavage events are conserved across metazoan model species. Using N-terminal labeling followed by mass spectrometry, we identified 257 caspase cleavage sites in mouse, 130 in Drosophila, and 50 in Caenorhabditis elegans. The large majority of the caspase cut sites identified in mouse proteins were found conserved in human orthologs. However, while many of the same proteins targeted in the more distantly related species were cleaved in human orthologs, the exact sites were often different. Furthermore, similar functional pathways are targeted by caspases in all four species. Our data suggest a model for the evolution of apoptotic caspase specificity that highlights the hierarchical importance of functional pathways over specific proteins, and proteins over their specific cleavage site motifs.
Figures
Similar articles
-
Novel Apoptotic Mediators Identified by Conservation of Vertebrate Caspase Targets.Biomolecules. 2020 Apr 15;10(4):612. doi: 10.3390/biom10040612. Biomolecules. 2020. PMID: 32326640 Free PMC article.
-
In Vitro Use of Peptide Based Substrates and Inhibitors of Apoptotic Caspases.Methods Mol Biol. 2016;1419:57-67. doi: 10.1007/978-1-4939-3581-9_5. Methods Mol Biol. 2016. PMID: 27108431
-
Cacidases: caspases can cleave after aspartate, glutamate and phosphoserine residues.Cell Death Differ. 2016 Oct;23(10):1717-26. doi: 10.1038/cdd.2016.62. Epub 2016 Jul 1. Cell Death Differ. 2016. PMID: 27367566 Free PMC article.
-
Caspases and their substrates.Cell Death Differ. 2017 Aug;24(8):1380-1389. doi: 10.1038/cdd.2017.44. Epub 2017 May 12. Cell Death Differ. 2017. PMID: 28498362 Free PMC article. Review.
-
Proteases for cell suicide: functions and regulation of caspases.Microbiol Mol Biol Rev. 2000 Dec;64(4):821-46. doi: 10.1128/MMBR.64.4.821-846.2000. Microbiol Mol Biol Rev. 2000. PMID: 11104820 Free PMC article. Review.
Cited by
-
Human ACAP2 is a homolog of C. elegans CNT-1 that promotes apoptosis in cancer cells.Cell Cycle. 2015;14(12):1771-8. doi: 10.1080/15384101.2015.1026518. Cell Cycle. 2015. PMID: 25853217 Free PMC article.
-
Circulating proteolytic signatures of chemotherapy-induced cell death in humans discovered by N-terminal labeling.Proc Natl Acad Sci U S A. 2014 May 27;111(21):7594-9. doi: 10.1073/pnas.1405987111. Epub 2014 May 12. Proc Natl Acad Sci U S A. 2014. PMID: 24821784 Free PMC article.
-
Caspase cleavage sites in the human proteome: CaspDB, a database of predicted substrates.PLoS One. 2014 Oct 17;9(10):e110539. doi: 10.1371/journal.pone.0110539. eCollection 2014. PLoS One. 2014. PMID: 25330111 Free PMC article.
-
Intrinsic apoptosis is evolutionarily divergent among metazoans.Evol Lett. 2023 Nov 16;8(2):267-282. doi: 10.1093/evlett/qrad057. eCollection 2024 Apr. Evol Lett. 2023. PMID: 38525035 Free PMC article.
-
Antioxidant Supplementation: A Linchpin in Radiation-Induced Enteritis.Technol Cancer Res Treat. 2017 Dec;16(6):676-691. doi: 10.1177/1533034617707598. Epub 2017 May 22. Technol Cancer Res Treat. 2017. PMID: 28532242 Free PMC article.
References
-
- Van Damme P, Martens L, Van Damme J, Hugelier K, Staes A, Vandekerckhove J, et al. Caspase-specific and nonspecific in vivo protein processing during Fas-induced apoptosis. Nat Methods. 2005;2:771–777. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

