Frequent amplification of CENPF, GMNN and CDK13 genes in hepatocellular carcinomas
- PMID: 22912832
- PMCID: PMC3418236
- DOI: 10.1371/journal.pone.0043223
Frequent amplification of CENPF, GMNN and CDK13 genes in hepatocellular carcinomas
Abstract
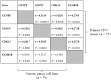
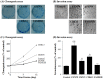
Genomic changes frequently occur in cancer cells during tumorigenesis from normal cells. Using the Illumina Human NS-12 single-nucleotide polymorphism (SNP) chip to screen for gene copy number changes in primary hepatocellular carcinomas (HCCs), we initially detected amplification of 35 genes from four genomic regions (1q21-41, 6p21.2-24.1, 7p13 and 8q13-23). By integrated screening of these genes for both DNA copy number and gene expression in HCC and colorectal cancer, we selected CENPF (centromere protein F/mitosin), GMNN (geminin, DNA replication inhibitor), CDK13 (cyclin-dependent kinase 13), and FAM82B (family with sequence similarity 82, member B) as common cancer genes. Each gene exhibited an amplification frequency of ~30% (range, 20-50%) in primary HCC (n = 57) and colorectal cancer (n = 12), as well as in a panel of human cancer cell lines (n = 70). Clonogenic and invasion assays of NIH3T3 cells transfected with each of the four amplified genes showed that CENPF, GMNN, and CDK13 were highly oncogenic whereas FAM82B was not. Interestingly, the oncogenic activity of these genes (excluding FAM82B) was highly correlated with gene-copy numbers in tumor samples (correlation coefficient, r>0.423), indicating that amplifications of CENPF, GMNN, and CDK13 genes are tightly linked and coincident in tumors. Furthermore, we confirmed that CDK13 gene copy number was significantly associated with clinical onset age in patients with HCC (P = 0.0037). Taken together, our results suggest that coincidently amplified CDK13, GMNN, and CENPF genes can play a role as common cancer-driver genes in human cancers.
Conflict of interest statement
Figures


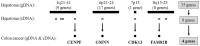


Similar articles
-
Centromere protein F promotes progression of hepatocellular carcinoma through ERK and cell cycle-associated pathways.Cancer Gene Ther. 2022 Jul;29(7):1033-1042. doi: 10.1038/s41417-021-00404-7. Epub 2021 Dec 2. Cancer Gene Ther. 2022. PMID: 34857915
-
Lymphoid-specific helicase promotes the growth and invasion of hepatocellular carcinoma by transcriptional regulation of centromere protein F expression.Cancer Sci. 2019 Jul;110(7):2133-2144. doi: 10.1111/cas.14037. Epub 2019 May 25. Cancer Sci. 2019. PMID: 31066149 Free PMC article.
-
Characterization of the oncogenic function of centromere protein F in hepatocellular carcinoma.Biochem Biophys Res Commun. 2013 Jul 12;436(4):711-8. doi: 10.1016/j.bbrc.2013.06.021. Epub 2013 Jun 17. Biochem Biophys Res Commun. 2013. PMID: 23791740
-
CDK1-PLK1/SGOL2/ANLN pathway mediating abnormal cell division in cell cycle may be a critical process in hepatocellular carcinoma.Cell Cycle. 2020 May;19(10):1236-1252. doi: 10.1080/15384101.2020.1749471. Epub 2020 Apr 10. Cell Cycle. 2020. PMID: 32275843 Free PMC article.
-
Geminin a multi task protein involved in cancer pathophysiology and developmental process: A review.Biochimie. 2016 Dec;131:115-127. doi: 10.1016/j.biochi.2016.09.022. Epub 2016 Oct 1. Biochimie. 2016. PMID: 27702582 Review.
Cited by
-
Identification of genes that are essential to restrict genome duplication to once per cell division.Oncotarget. 2016 Jun 7;7(23):34956-76. doi: 10.18632/oncotarget.9008. Oncotarget. 2016. PMID: 27144335 Free PMC article.
-
Loss of CENPF leads to developmental failure in mouse embryos.Cell Cycle. 2019 Oct;18(20):2784-2799. doi: 10.1080/15384101.2019.1661173. Epub 2019 Sep 3. Cell Cycle. 2019. PMID: 31478449 Free PMC article.
-
Silencing proline-rich coiled-coil 2C inhibit the proliferation and metastasis of liver cancer cells.J Gastrointest Oncol. 2023 Feb 28;14(1):287-301. doi: 10.21037/jgo-23-10. J Gastrointest Oncol. 2023. PMID: 36915448 Free PMC article.
-
MiR-1-3p targets CENPF to repress tumor-relevant functions of gastric cancer cells.BMC Gastroenterol. 2022 Mar 28;22(1):145. doi: 10.1186/s12876-022-02203-2. BMC Gastroenterol. 2022. PMID: 35346060 Free PMC article.
-
Mouse Model of Congenital Heart Defects, Dysmorphic Facial Features and Intellectual Developmental Disorders as a Result of Non-functional CDK13.Front Cell Dev Biol. 2019 Aug 7;7:155. doi: 10.3389/fcell.2019.00155. eCollection 2019. Front Cell Dev Biol. 2019. PMID: 31440507 Free PMC article.
References
-
- Albertson DG, Collins C, McCormick F, Gray JW (2003) Chromosomal aberrations in solid tumors. Nature Genet 34: 369–376. - PubMed
-
- Medina PP, Castillo SD, Blanco S, Sanz-Garcia M, Largo C, et al. (2009) The SRY-HMG box gene, SOX4, is a target of gene amplification at chromosome 6p in lung cancer. Hum Mol Genet 18: 1343–1352. - PubMed
-
- Liu W, Chang B, Sauvageot J, Dimitrov L, Gielzak M, et al. (2006) Comprehensive assessment of DNA copy number alterations in human prostate cancers using Affymetrix 100K SNP mapping array. Genes Chromosomes Cancer 45: 1018–1032. - PubMed
-
- Santarius T, Shipley J, Brewer D, Stratton MR, Cooper CS (2010) A census of amplified and overexpressed human cancer genes. Nature Rev Cancer 10: 59–64. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

