Loss of a 20S proteasome activator in Saccharomyces cerevisiae downregulates genes important for genomic integrity, increases DNA damage, and selectively sensitizes cells to agents with diverse mechanisms of action
- PMID: 22908043
- PMCID: PMC3411250
- DOI: 10.1534/g3.112.003376
Loss of a 20S proteasome activator in Saccharomyces cerevisiae downregulates genes important for genomic integrity, increases DNA damage, and selectively sensitizes cells to agents with diverse mechanisms of action
Abstract
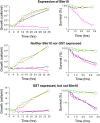
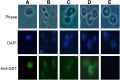
Cytoprotective functions of a 20S proteasome activator were investigated. Saccharomyces cerevisiae Blm10 and human 20S proteasome activator 200 (PA200) are homologs. Comparative genome-wide analyses of untreated diploid cells lacking Blm10 and growing at steady state at defined growth rates revealed downregulation of numerous genes required for accurate chromosome structure, assembly and repair, and upregulation of a specific subset of genes encoding protein-folding chaperones. Blm10 loss or truncation of the Ubp3/Blm3 deubiquitinating enzyme caused massive chromosomal damage and cell death in homozygous diploids after phleomycin treatments, indicating that Blm10 and Ubp3/Blm3 function to stabilize the genome and protect against cell death. Diploids lacking Blm10 also were sensitized to doxorubicin, hydroxyurea, 5-fluorouracil, rapamycin, hydrogen peroxide, methyl methanesulfonate, and calcofluor. Fluorescently tagged Blm10 localized in nuclei, with enhanced fluorescence after DNA replication. After DNA damage that caused a classic G2/M arrest, fluorescence remained diffuse, with evidence of nuclear fragmentation in some cells. Protective functions of Blm10 did not require the carboxyl-terminal region that makes close contact with 20S proteasomes, indicating that protection does not require this contact or the truncated Blm10 can interact with the proteasome apart from this region. Without its carboxyl-terminus, Blm10((-339aa)) localized to nuclei in untreated, nonproliferating (G(0)) cells, but not during G(1) S, G(2), and M. The results indicate Blm10 functions in protective mechanisms that include the machinery that assures proper assembly of chromosomes. These essential guardian functions have implications for ubiquitin-independent targeting in anticancer therapy. Targeting Blm10/PA200 together with one or more of the upregulated chaperones or a conventional treatment could be efficacious.
Keywords: 20S proteasome activator; BLM10/PA200; DNA damage; UBP3/BLM3; molecular chaperones.
Figures



Similar articles
-
blm3-1 is an allele of UBP3, a ubiquitin protease that appears to act during transcription of damaged DNA.J Mol Biol. 2006 Oct 27;363(3):660-72. doi: 10.1016/j.jmb.2006.08.073. Epub 2006 Aug 30. J Mol Biol. 2006. PMID: 16997324
-
The Proteasome Activators Blm10/PA200 Enhance the Proteasomal Degradation of N-Terminal Huntingtin.Biomolecules. 2020 Nov 20;10(11):1581. doi: 10.3390/biom10111581. Biomolecules. 2020. PMID: 33233776 Free PMC article.
-
Structure of a Blm10 complex reveals common mechanisms for proteasome binding and gate opening.Mol Cell. 2010 Mar 12;37(5):728-35. doi: 10.1016/j.molcel.2010.02.002. Mol Cell. 2010. PMID: 20227375 Free PMC article.
-
Proteasome activator 200: the heat is on..Mol Cell Proteomics. 2011 May;10(5):R110.006890. doi: 10.1074/mcp.R110.006890. Epub 2011 Mar 9. Mol Cell Proteomics. 2011. PMID: 21389348 Free PMC article. Review.
-
PA200-Mediated Proteasomal Protein Degradation and Regulation of Cellular Senescence.Int J Mol Sci. 2024 May 22;25(11):5637. doi: 10.3390/ijms25115637. Int J Mol Sci. 2024. PMID: 38891826 Free PMC article. Review.
Cited by
-
The proteasome activator PA200 regulates expression of genes involved in cell survival upon selective mitochondrial inhibition in neuroblastoma cells.J Cell Mol Med. 2020 Jun;24(12):6716-6730. doi: 10.1111/jcmm.15323. Epub 2020 May 5. J Cell Mol Med. 2020. PMID: 32368861 Free PMC article.
-
Nuclear Transport of Yeast Proteasomes.Front Mol Biosci. 2019 May 16;6:34. doi: 10.3389/fmolb.2019.00034. eCollection 2019. Front Mol Biosci. 2019. PMID: 31157235 Free PMC article. Review.
-
Chromatin stiffening underlies enhanced locus mobility after DNA damage in budding yeast.EMBO J. 2017 Sep 1;36(17):2595-2608. doi: 10.15252/embj.201695842. Epub 2017 Jul 10. EMBO J. 2017. PMID: 28694242 Free PMC article.
-
Regulation of proteasome activity in health and disease.Biochim Biophys Acta. 2014 Jan;1843(1):13-25. doi: 10.1016/j.bbamcr.2013.08.012. Epub 2013 Aug 27. Biochim Biophys Acta. 2014. PMID: 23994620 Free PMC article. Review.
-
Histone degradation by the proteasome regulates chromatin and cellular plasticity.FEBS J. 2022 Jun;289(12):3304-3316. doi: 10.1111/febs.15903. Epub 2021 May 13. FEBS J. 2022. PMID: 33914417 Free PMC article. Review.
References
-
- Aouida M., Page N., Leduc A., Peter M., Ramotar D., 2004. A genome-wide screen in Saccharomyces cerevisiae reveals altered transport as a mechanism of resistance to the anticancer drug bleomycin. Cancer Res. 64: 1102–1109 - PubMed
-
- Awasthi N., Wagner B. J., 2005. Upregulation of heat shock protein expression by proteasome inhibition: an antiapoptotic mechanism in the lens. Invest. Ophthalmol. Vis. Sci. 46: 2082–2091 - PubMed
-
- Bachmair A., Finley D., Varshavsky A., 1986. In vivo half-life of a protein is a function of its amino-terminal residue. Science 234: 179–186 - PubMed
-
- Baxter B. K., Craig E. A., 1998. Isolation of UBP3, encoding a de-ubiquitinating enzyme, as a multicopy suppressor of a heat-shock mutant strain of S. cerevisiae. Curr. Genet. 33: 412–419 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
