PKA isoforms coordinate mRNA fate during nutrient starvation
- PMID: 22899713
- PMCID: PMC3533396
- DOI: 10.1242/jcs.111534
PKA isoforms coordinate mRNA fate during nutrient starvation
Abstract
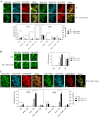
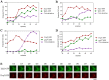
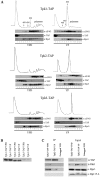
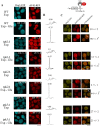
A variety of stress conditions induce mRNA and protein aggregation into mRNA silencing foci, but the signalling pathways mediating these responses are still elusive. Previously we demonstrated that PKA catalytic isoforms Tpk2 and Tpk3 localise with processing and stress bodies in Saccharomyces cerevisiae. Here, we show that Tpk2 and Tpk3 are associated with translation initiation factors Pab1 and Rps3 in exponentially growing cells. Glucose starvation promotes the loss of interaction between Tpk and initiation factors followed by their accumulation into processing bodies. Analysis of mutants of the individual PKA isoform genes has revealed that the TPK3 or TPK2 deletion affects the capacity of the cells to form granules and arrest translation properly in response to glucose starvation or stationary phase. Moreover, we demonstrate that PKA controls Rpg1 and eIF4G(1) protein abundance, possibly controlling cap-dependent translation. Taken together, our data suggest that the PKA pathway coordinates multiple stages in the fate of mRNAs in association with nutritional environment and growth status of the cell.
Figures


Similar articles
-
The role of PKA in the translational response to heat stress in Saccharomyces cerevisiae.PLoS One. 2017 Oct 18;12(10):e0185416. doi: 10.1371/journal.pone.0185416. eCollection 2017. PLoS One. 2017. PMID: 29045428 Free PMC article.
-
Differential localization to cytoplasm, nucleus or P-bodies of yeast PKA subunits under different growth conditions.Eur J Cell Biol. 2010 Apr;89(4):339-48. doi: 10.1016/j.ejcb.2009.08.005. Epub 2009 Oct 4. Eur J Cell Biol. 2010. PMID: 19804918
-
The activation loop of PKA catalytic isoforms is differentially phosphorylated by Pkh protein kinases in Saccharomyces cerevisiae.Biochem J. 2012 Dec 15;448(3):307-20. doi: 10.1042/BJ20121061. Biochem J. 2012. PMID: 22957732
-
eIF4G-an integrator of mRNA metabolism?FEMS Yeast Res. 2016 Nov;16(7):fow087. doi: 10.1093/femsyr/fow087. Epub 2016 Sep 29. FEMS Yeast Res. 2016. PMID: 27694156 Review.
-
The Saccharomyces cerevisiae poly (A) binding protein (Pab1): Master regulator of mRNA metabolism and cell physiology.Yeast. 2019 Jan;36(1):23-34. doi: 10.1002/yea.3347. Epub 2018 Oct 17. Yeast. 2019. PMID: 30006991 Review.
Cited by
-
Granules harboring translationally active mRNAs provide a platform for P-body formation following stress.Cell Rep. 2014 Nov 6;9(3):944-54. doi: 10.1016/j.celrep.2014.09.040. Epub 2014 Oct 23. Cell Rep. 2014. PMID: 25437551 Free PMC article.
-
Light-sensing via hydrogen peroxide and a peroxiredoxin.Nat Commun. 2017 Mar 24;8:14791. doi: 10.1038/ncomms14791. Nat Commun. 2017. PMID: 28337980 Free PMC article.
-
Yeast Secretes High Amounts of Human Calreticulin without Cellular Stress.Curr Issues Mol Biol. 2022 Apr 19;44(5):1768-1787. doi: 10.3390/cimb44050122. Curr Issues Mol Biol. 2022. PMID: 35678651 Free PMC article.
-
The role of PKA in the translational response to heat stress in Saccharomyces cerevisiae.PLoS One. 2017 Oct 18;12(10):e0185416. doi: 10.1371/journal.pone.0185416. eCollection 2017. PLoS One. 2017. PMID: 29045428 Free PMC article.
-
Mechanism and Regulation of Protein Synthesis in Saccharomyces cerevisiae.Genetics. 2016 May;203(1):65-107. doi: 10.1534/genetics.115.186221. Genetics. 2016. PMID: 27183566 Free PMC article. Review.
References
-
- Beullens M., Mbonyi K., Geerts L., Gladines D., Detremerie K., Jans A. W., Thevelein J. M. (1988). Studies on the mechanism of the glucose-induced cAMP signal in glycolysis and glucose repression mutants of the yeast Saccharomyces cerevisiae. Eur. J. Biochem. 172, 227–231 10.1111/j.1432-1033.1988.tb13877.x - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

