Pseudogenization of the MCP-2/CCL8 chemokine gene in European rabbit (genus Oryctolagus), but not in species of Cottontail rabbit (Sylvilagus) and Hare (Lepus)
- PMID: 22894773
- PMCID: PMC3511233
- DOI: 10.1186/1471-2156-13-72
Pseudogenization of the MCP-2/CCL8 chemokine gene in European rabbit (genus Oryctolagus), but not in species of Cottontail rabbit (Sylvilagus) and Hare (Lepus)
Abstract
Background: Recent studies in human have highlighted the importance of the monocyte chemotactic proteins (MCP) in leukocyte trafficking and their effects in inflammatory processes, tumor progression, and HIV-1 infection. In European rabbit (Oryctolagus cuniculus) one of the prime MCP targets, the chemokine receptor CCR5 underwent a unique structural alteration. Until now, no homologue of MCP-2/CCL8a, MCP-3/CCL7 or MCP-4/CCL13 genes have been reported for this species. This is interesting, because at least the first two genes are expressed in most, if not all, mammals studied, and appear to be implicated in a variety of important chemokine ligand-receptor interactions. By assessing the Rabbit Whole Genome Sequence (WGS) data we have searched for orthologs of the mammalian genes of the MCP-Eotaxin cluster.
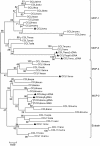
Results: We have localized the orthologs of these chemokine genes in the genome of European rabbit and compared them to those of leporid genera which do (i.e. Oryctolagus and Bunolagus) or do not share the CCR5 alteration with European rabbit (i.e. Lepus and Sylvilagus). Of the Rabbit orthologs of the CCL8, CCL7, and CCL13 genes only the last two were potentially functional, although showing some structural anomalies at the protein level. The ortholog of MCP-2/CCL8 appeared to be pseudogenized by deleterious nucleotide substitutions affecting exon1 and exon2. By analyzing both genomic and cDNA products, these studies were extended to wild specimens of four genera of the Leporidae family: Oryctolagus, Bunolagus, Lepus, and Sylvilagus. It appeared that the anomalies of the MCP-3/CCL7 and MCP-4/CCL13 proteins are shared among the different species of leporids. In contrast, whereas MCP-2/CCL8 was pseudogenized in every studied specimen of the Oryctolagus - Bunolagus lineage, this gene was intact in species of the Lepus - Sylvilagus lineage, and was, at least in Lepus, correctly transcribed.
Conclusion: The biological function of a gene was often revealed in situations of dysfunction or gene loss. Infections with Myxoma virus (MYXV) tend to be fatal in European rabbit (genus Oryctolagus), while being harmless in Hares (genus Lepus) and benign in Cottontail rabbit (genus Sylvilagus), the natural hosts of the virus. This communication should stimulate research on a possible role of MCP-2/CCL8 in poxvirus related pathogenicity.
Figures

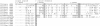




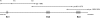
Similar articles
-
Evolution of viral sensing RIG-I-like receptor genes in Leporidae genera Oryctolagus, Sylvilagus, and Lepus.Immunogenetics. 2014 Jan;66(1):43-52. doi: 10.1007/s00251-013-0740-7. Epub 2013 Nov 13. Immunogenetics. 2014. PMID: 24220721
-
Genetic variation at chemokine receptor CCR5 in leporids: alteration at the 2nd extracellular domain by gene conversion with CCR2 in Oryctolagus, but not in Sylvilagus and Lepus species.Immunogenetics. 2006 Jun;58(5-6):494-501. doi: 10.1007/s00251-006-0095-4. Epub 2006 Apr 5. Immunogenetics. 2006. PMID: 16596402
-
Study of Sylvilagus rabbit TRIM5α species-specific domain: how ancient endoviruses could have shaped the antiviral repertoire in Lagomorpha.BMC Evol Biol. 2011 Oct 8;11:294. doi: 10.1186/1471-2148-11-294. BMC Evol Biol. 2011. PMID: 21982459 Free PMC article.
-
Rabbit hemorrhagic disease virus 2, 2010-2023: a review of global detections and affected species.J Vet Diagn Invest. 2024 Sep;36(5):617-637. doi: 10.1177/10406387241260281. Epub 2024 Jun 19. J Vet Diagn Invest. 2024. PMID: 39344909 Review.
-
Human monocyte chemotactic proteins-2 and -3: structural and functional comparison with MCP-1.J Leukoc Biol. 1996 Jan;59(1):67-74. doi: 10.1002/jlb.59.1.67. J Leukoc Biol. 1996. PMID: 8558070 Review.
Cited by
-
Evolution of CCL16 in Glires (Rodentia and Lagomorpha) shows an unusual random pseudogenization pattern.BMC Evol Biol. 2019 Feb 20;19(1):59. doi: 10.1186/s12862-019-1390-7. BMC Evol Biol. 2019. PMID: 30786851 Free PMC article.
-
An intriguing shift occurs in the novel protein phosphatase 1 binding partner, TCTEX1D4: evidence of positive selection in a pika model.PLoS One. 2013 Oct 10;8(10):e77236. doi: 10.1371/journal.pone.0077236. eCollection 2013. PLoS One. 2013. PMID: 24130861 Free PMC article.
-
Adaptive Gene Loss? Tracing Back the Pseudogenization of the Rabbit CCL8 Chemokine.J Mol Evol. 2016 Aug;83(1-2):12-25. doi: 10.1007/s00239-016-9747-7. Epub 2016 Jun 15. J Mol Evol. 2016. PMID: 27306379
-
What Are the Keys to the Adaptive Success of European Wild Rabbit (Oryctolagus cuniculus) in the Iberian Peninsula?Animals (Basel). 2021 Aug 20;11(8):2453. doi: 10.3390/ani11082453. Animals (Basel). 2021. PMID: 34438909 Free PMC article. Review.
-
Convergent inactivation of the skin-specific C-C motif chemokine ligand 27 in mammalian evolution.Immunogenetics. 2019 May;71(5-6):363-372. doi: 10.1007/s00251-019-01114-z. Epub 2019 May 2. Immunogenetics. 2019. PMID: 31049641
References
-
- Lopez-Martinez N. Revision sistematica y biostratigrafica de los lagomorphos (Mammalia) del neogeno y cuaternario de España. Disputacion General de Aragon: Memorias del Museo Paleontologico de la Universidad de Zaragoza; 1989.
-
- Chapman JA, Flux JEC. Rabbits, hares and pikas: status survey and conservation action plan. IUCN/SSC Lagomorph Specialist Group (ed.) Oxford UK: Information Press; 1990.
-
- Rougeot J. Origin et histoire du lapin. Ethnozootechnie. 1981;27:1–9.
-
- Flux JEC. In: The European rabbit: The history of a successful colonizer. Thomson HV, King CM, editor. Oxford: Oxford Science Publications; 1994. World distribution; pp. 8–21.
-
- Callou C. Modifications de l'aire de répartition du Lapin (Oryctolagus cuniculus) en France et en Espagne, du Pléistocène à l'époque actuelle État de la question. Anthropozoologica. 1995;21:95–114.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

