Human intron-encoded Alu RNAs are processed and packaged into Wdr79-associated nucleoplasmic box H/ACA RNPs
- PMID: 22892240
- PMCID: PMC3435494
- DOI: 10.1101/gad.197467.112
Human intron-encoded Alu RNAs are processed and packaged into Wdr79-associated nucleoplasmic box H/ACA RNPs
Abstract
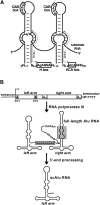
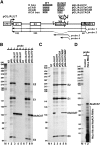
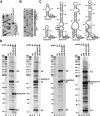
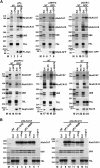
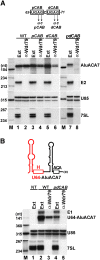
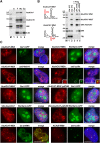
Alu repetitive sequences are the most abundant short interspersed DNA elements in the human genome. Full-length Alu elements are composed of two tandem sequence monomers, the left and right Alu arms, both derived from the 7SL signal recognition particle RNA. Since Alu elements are common in protein-coding genes, they are frequently transcribed into pre-mRNAs. Here, we demonstrate that the right arms of nascent Alu transcripts synthesized within pre-mRNA introns are processed into metabolically stable small RNAs. The intron-encoded Alu RNAs, termed AluACA RNAs, are structurally highly reminiscent of box H/ACA small Cajal body (CB) RNAs (scaRNAs). They are composed of two hairpin units followed by the essential H (AnAnnA) and ACA box motifs. The mature AluACA RNAs associate with the four H/ACA core proteins: dyskerin, Nop10, Nhp2, and Gar1. Moreover, the 3' hairpin of AluACA RNAs carries two closely spaced CB localization motifs, CAB boxes (UGAG), which bind Wdr79 in a cumulative fashion. In contrast to canonical H/ACA scaRNPs, which concentrate in CBs, the AluACA RNPs accumulate in the nucleoplasm. Identification of 348 human AluACA RNAs demonstrates that intron-encoded AluACA RNAs represent a novel, large subgroup of H/ACA RNAs, which are apparently confined to human or primate cells.
Figures

Similar articles
-
Human intron-encoded AluACA RNAs and telomerase RNA share a common element promoting RNA accumulation.RNA Biol. 2016 Dec;13(12):1274-1285. doi: 10.1080/15476286.2016.1239689. Epub 2016 Oct 11. RNA Biol. 2016. PMID: 27726486 Free PMC article.
-
Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs.Cold Spring Harb Symp Quant Biol. 2006;71:407-17. doi: 10.1101/sqb.2006.71.025. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381323 Review.
-
Targeting vertebrate intron-encoded box C/D 2'-O-methylation guide RNAs into the Cajal body.Nucleic Acids Res. 2014 Jun;42(10):6616-29. doi: 10.1093/nar/gku287. Epub 2014 Apr 20. Nucleic Acids Res. 2014. PMID: 24753405 Free PMC article.
-
Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins.EMBO J. 2004 Apr 21;23(8):1857-67. doi: 10.1038/sj.emboj.7600181. Epub 2004 Mar 25. EMBO J. 2004. PMID: 15044956 Free PMC article.
-
RNA structure and function in C/D and H/ACA s(no)RNPs.Curr Opin Struct Biol. 2004 Jun;14(3):335-43. doi: 10.1016/j.sbi.2004.05.006. Curr Opin Struct Biol. 2004. PMID: 15193314 Review.
Cited by
-
WDR79 promotes aerobic glycolysis of pancreatic ductal adenocarcinoma (PDAC) by the suppression of SIRT4.Open Med (Wars). 2023 Jan 16;18(1):20220624. doi: 10.1515/med-2022-0624. eCollection 2023. Open Med (Wars). 2023. PMID: 36712589 Free PMC article.
-
Cooperative 2'-O-methylation of the wobble cytidine of human elongator tRNAMet(CAT) by a nucleolar and a Cajal body-specific box C/D RNP.Genes Dev. 2019 Jul 1;33(13-14):741-746. doi: 10.1101/gad.326363.119. Epub 2019 Jun 6. Genes Dev. 2019. PMID: 31171702 Free PMC article.
-
Small RNAs with big implications: new insights into H/ACA snoRNA function and their role in human disease.Wiley Interdiscip Rev RNA. 2015 Mar-Apr;6(2):173-89. doi: 10.1002/wrna.1266. Epub 2014 Oct 31. Wiley Interdiscip Rev RNA. 2015. PMID: 25363811 Free PMC article. Review.
-
Pearls are novel Cajal body-like structures in the Xenopus germinal vesicle that are dependent on RNA pol III transcription.Chromosome Res. 2012 Dec;20(8):953-69. doi: 10.1007/s10577-012-9320-1. Chromosome Res. 2012. PMID: 23135638 Free PMC article.
-
RNA pseudouridylation: new insights into an old modification.Trends Biochem Sci. 2013 Apr;38(4):210-8. doi: 10.1016/j.tibs.2013.01.002. Epub 2013 Feb 4. Trends Biochem Sci. 2013. PMID: 23391857 Free PMC article. Review.
References
-
- Balakin AG, Smith L, Fournier MJ 1996. The RNA world of the nucleolus: Two major families of small RNAs defined by different box elements with related functions. Cell 86: 823–834 - PubMed
-
- Batzer MA, Deininger PL 2002. Alu repeats and human genomic diversity. Nat Rev Genet 3: 370–379 - PubMed
-
- Berger A, Strub K 2011. Multiple roles of Alu-related noncoding RNAs. Prog Mol Subcell Biol 51: 119–146 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
