Mutations to the formin homology 2 domain of INF2 protein have unexpected effects on actin polymerization and severing
- PMID: 22879592
- PMCID: PMC3464531
- DOI: 10.1074/jbc.M112.365122
Mutations to the formin homology 2 domain of INF2 protein have unexpected effects on actin polymerization and severing
Abstract
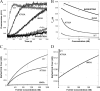
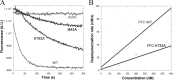
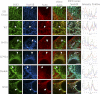
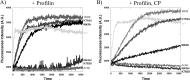
INF2 (inverted formin 2) is a formin protein with unusual biochemical characteristics. As with other formins, the formin homology 2 (FH2) domain of INF2 accelerates actin filament assembly and remains at the barbed end, modulating elongation. The unique feature of INF2 is its ability to sever filaments and enhance depolymerization, which requires the C-terminal region. Physiologically, INF2 acts in the secretory pathway and is mutated in two human diseases, focal and segmental glomerulosclerosis and Charcot-Marie-Tooth disease. In this study, we investigate the effects of mutating two FH2 residues found to be key in other formins: Ile-643 and Lys-792. Surprisingly, neither mutation abolishes barbed end binding, as judged by pyrene-actin and total internal reflection (TIRF) microscopy elongation assays. The I643A mutation causes tight capping of a subset of filaments, whereas K792A causes slow elongation of all filaments. The I643A mutation has a minor inhibitory effect on polymerization activity but causes almost complete abolition of severing and depolymerization activity. The K792A mutation has relatively small effects on polymerization, severing, and depolymerization. In cells, the K792A mutant causes actin accumulation around the endoplasmic reticulum to a similar extent as wild type, whereas the I643A mutant causes no measurable polymerization. The inability of I643A to induce actin polymerization in cells is explained by its inability to promote robust actin polymerization in the presence of capping protein. These results highlight an important point: it is dangerous to assume that mutation of conserved FH2 residues will have equivalent effects in all formins. The work also suggests that both mutations have effects on the mechanism of processive elongation.
Figures



Similar articles
-
INF2 Is a WASP homology 2 motif-containing formin that severs actin filaments and accelerates both polymerization and depolymerization.J Biol Chem. 2006 Sep 8;281(36):26754-67. doi: 10.1074/jbc.M604666200. Epub 2006 Jul 3. J Biol Chem. 2006. PMID: 16818491
-
The C terminus of formin FMNL3 accelerates actin polymerization and contains a WH2 domain-like sequence that binds both monomers and filament barbed ends.J Biol Chem. 2012 Jan 27;287(5):3087-98. doi: 10.1074/jbc.M111.312207. Epub 2011 Nov 17. J Biol Chem. 2012. PMID: 22094460 Free PMC article.
-
Actin monomers activate inverted formin 2 by competing with its autoinhibitory interaction.J Biol Chem. 2013 Sep 13;288(37):26847-55. doi: 10.1074/jbc.M113.472415. Epub 2013 Aug 6. J Biol Chem. 2013. PMID: 23921379 Free PMC article.
-
Role of formin INF2 in human diseases.Mol Biol Rep. 2022 Jan;49(1):735-746. doi: 10.1007/s11033-021-06869-x. Epub 2021 Oct 26. Mol Biol Rep. 2022. PMID: 34698992 Review.
-
Review of the mechanism of processive actin filament elongation by formins.Cell Motil Cytoskeleton. 2009 Aug;66(8):606-17. doi: 10.1002/cm.20379. Cell Motil Cytoskeleton. 2009. PMID: 19459187 Free PMC article. Review.
Cited by
-
The activities of the C-terminal regions of the formin protein disheveled-associated activator of morphogenesis (DAAM) in actin dynamics.J Biol Chem. 2017 Aug 18;292(33):13566-13583. doi: 10.1074/jbc.M117.799247. Epub 2017 Jun 22. J Biol Chem. 2017. PMID: 28642367 Free PMC article.
-
Assembly and turnover of short actin filaments by the formin INF2 and profilin.J Biol Chem. 2015 Sep 11;290(37):22494-506. doi: 10.1074/jbc.M115.670166. Epub 2015 Jun 29. J Biol Chem. 2015. PMID: 26124273 Free PMC article.
-
An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2.Science. 2013 Jan 25;339(6118):464-7. doi: 10.1126/science.1228360. Science. 2013. PMID: 23349293 Free PMC article.
-
The neuron-specific formin Delphilin nucleates nonmuscle actin but does not enhance elongation.Mol Biol Cell. 2018 Mar 1;29(5):610-621. doi: 10.1091/mbc.E17-06-0363. Epub 2017 Dec 27. Mol Biol Cell. 2018. PMID: 29282276 Free PMC article.
-
FMNL2 and -3 regulate Golgi architecture and anterograde transport downstream of Cdc42.Sci Rep. 2017 Aug 29;7(1):9791. doi: 10.1038/s41598-017-09952-1. Sci Rep. 2017. PMID: 28852060 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

