Structural bases of coronavirus attachment to host aminopeptidase N and its inhibition by neutralizing antibodies
- PMID: 22876187
- PMCID: PMC3410853
- DOI: 10.1371/journal.ppat.1002859
Structural bases of coronavirus attachment to host aminopeptidase N and its inhibition by neutralizing antibodies
Abstract
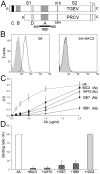
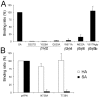
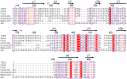
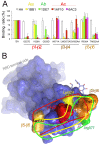
The coronaviruses (CoVs) are enveloped viruses of animals and humans associated mostly with enteric and respiratory diseases, such as the severe acute respiratory syndrome and 10-20% of all common colds. A subset of CoVs uses the cell surface aminopeptidase N (APN), a membrane-bound metalloprotease, as a cell entry receptor. In these viruses, the envelope spike glycoprotein (S) mediates the attachment of the virus particles to APN and subsequent cell entry, which can be blocked by neutralizing antibodies. Here we describe the crystal structures of the receptor-binding domains (RBDs) of two closely related CoV strains, transmissible gastroenteritis virus (TGEV) and porcine respiratory CoV (PRCV), in complex with their receptor, porcine APN (pAPN), or with a neutralizing antibody. The data provide detailed information on the architecture of the dimeric pAPN ectodomain and its interaction with the CoV S. We show that a protruding receptor-binding edge in the S determines virus-binding specificity for recessed glycan-containing surfaces in the membrane-distal region of the pAPN ectodomain. Comparison of the RBDs of TGEV and PRCV to those of other related CoVs, suggests that the conformation of the S receptor-binding region determines cell entry receptor specificity. Moreover, the receptor-binding edge is a major antigenic determinant in the TGEV envelope S that is targeted by neutralizing antibodies. Our results provide a compelling view on CoV cell entry and immune neutralization, and may aid the design of antivirals or CoV vaccines. APN is also considered a target for cancer therapy and its structure, reported here, could facilitate the development of anti-cancer drugs.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Porcine Deltacoronavirus Engages the Transmissible Gastroenteritis Virus Functional Receptor Porcine Aminopeptidase N for Infectious Cellular Entry.J Virol. 2018 May 29;92(12):e00318-18. doi: 10.1128/JVI.00318-18. Print 2018 Jun 15. J Virol. 2018. PMID: 29618640 Free PMC article.
-
Further characterization of aminopeptidase-N as a receptor for coronaviruses.Adv Exp Med Biol. 1993;342:293-8. doi: 10.1007/978-1-4615-2996-5_45. Adv Exp Med Biol. 1993. PMID: 7911642
-
Recombinant Receptor-Binding Domains of Multiple Middle East Respiratory Syndrome Coronaviruses (MERS-CoVs) Induce Cross-Neutralizing Antibodies against Divergent Human and Camel MERS-CoVs and Antibody Escape Mutants.J Virol. 2016 Dec 16;91(1):e01651-16. doi: 10.1128/JVI.01651-16. Print 2017 Jan 1. J Virol. 2016. PMID: 27795425 Free PMC article.
-
Neutralizing ebolavirus: structural insights into the envelope glycoprotein and antibodies targeted against it.Curr Opin Struct Biol. 2009 Aug;19(4):408-17. doi: 10.1016/j.sbi.2009.05.004. Epub 2009 Jun 24. Curr Opin Struct Biol. 2009. PMID: 19559599 Free PMC article. Review.
-
Structural basis for differential neutralization of ebolaviruses.Viruses. 2012 Apr;4(4):447-70. doi: 10.3390/v4040447. Epub 2012 Apr 5. Viruses. 2012. PMID: 22590681 Free PMC article. Review.
Cited by
-
The virus-host interface: Molecular interactions of Alphacoronavirus-1 variants from wild and domestic hosts with mammalian aminopeptidase N.Mol Ecol. 2021 Jun;30(11):2607-2625. doi: 10.1111/mec.15910. Epub 2021 May 3. Mol Ecol. 2021. PMID: 33786949 Free PMC article.
-
Genotyping coronaviruses associated with feline infectious peritonitis.J Gen Virol. 2015 Jun;96(Pt 6):1358-1368. doi: 10.1099/vir.0.000084. Epub 2015 Feb 9. J Gen Virol. 2015. PMID: 25667330 Free PMC article.
-
SARS-CoV-2 cell entry beyond the ACE2 receptor.Mol Biol Rep. 2022 Nov;49(11):10715-10727. doi: 10.1007/s11033-022-07700-x. Epub 2022 Jun 26. Mol Biol Rep. 2022. PMID: 35754059 Free PMC article. Review.
-
Identification of a Novel Neutralizing Epitope on the N-Terminal Domain of the Human Coronavirus 229E Spike Protein.J Virol. 2022 Feb 23;96(4):e0195521. doi: 10.1128/JVI.01955-21. Epub 2021 Dec 15. J Virol. 2022. PMID: 34908442 Free PMC article.
-
Receptor recognition mechanisms of coronaviruses: a decade of structural studies.J Virol. 2015 Feb;89(4):1954-64. doi: 10.1128/JVI.02615-14. Epub 2014 Nov 26. J Virol. 2015. PMID: 25428871 Free PMC article. Review.
References
-
- Enjuanes L, Gorbalenya AE, de Groot RJ, Cowley JA, Ziebuhr J, et al. (2008) Nidovirales. In: Encyclopedia of Virology. Third ed. Mahy BWJ, Van Regenmortel MHV, editors. Oxford: Elsevier. 419–430.
-
- de Groot RJ, Baker SC, Baric R, Enjuanes L, Gorbalenya AE, et al. (2011) Coronaviridae. In: Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. San Diego: Elsevier Academic Press. 774–796.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

