Distinct loops in arrestin differentially regulate ligand binding within the GPCR opsin
- PMID: 22871814
- PMCID: PMC3455371
- DOI: 10.1038/ncomms2000
Distinct loops in arrestin differentially regulate ligand binding within the GPCR opsin
Erratum in
- Nat Commun. 2012;3:1273
Abstract
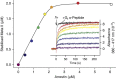
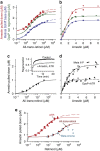
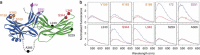
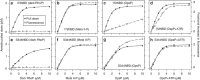
G-protein-coupled receptors are universally regulated by arrestin binding. Here we show that rod arrestin induces uptake of the agonist all-trans-retinal [corrected] in only half the population of phosphorylated opsin in the native membrane. Agonist uptake blocks subsequent entry of the inverse agonist 11-cis-retinal (that is, regeneration of rhodopsin), but regeneration is not blocked in the other half of aporeceptors. Environmentally sensitive fluorophores attached to arrestin reported that conformational changes in loop(V-VI) (N-domain) are coupled to the entry of agonist, while loop(XVIII-XIX) (C-domain) engages the aporeceptor even before agonist is added. The data are most consistent with a model in which each domain of arrestin engages its own aporeceptor, and the different binding preferences of the domains lead to asymmetric ligand binding by the aporeceptors. Such a mechanism would protect the rod cell in bright light by concurrently sequestering toxic all-trans-retinal [corrected] and allowing regeneration with 11-cis-retinal.
Figures


Similar articles
-
Formation and decay of the arrestin·rhodopsin complex in native disc membranes.J Biol Chem. 2015 May 15;290(20):12919-28. doi: 10.1074/jbc.M114.620898. Epub 2015 Apr 6. J Biol Chem. 2015. PMID: 25847250 Free PMC article.
-
Conformational selection and equilibrium governs the ability of retinals to bind opsin.J Biol Chem. 2015 Feb 13;290(7):4304-18. doi: 10.1074/jbc.M114.603134. Epub 2014 Dec 1. J Biol Chem. 2015. PMID: 25451936 Free PMC article.
-
Functional map of arrestin binding to phosphorylated opsin, with and without agonist.Sci Rep. 2016 Jun 28;6:28686. doi: 10.1038/srep28686. Sci Rep. 2016. PMID: 27350090 Free PMC article.
-
The structural basis of the arrestin binding to GPCRs.Mol Cell Endocrinol. 2019 Mar 15;484:34-41. doi: 10.1016/j.mce.2019.01.019. Epub 2019 Jan 28. Mol Cell Endocrinol. 2019. PMID: 30703488 Free PMC article. Review.
-
Relevance of rhodopsin studies for GPCR activation.Biochim Biophys Acta. 2014 May;1837(5):674-82. doi: 10.1016/j.bbabio.2013.09.002. Epub 2013 Sep 13. Biochim Biophys Acta. 2014. PMID: 24041646 Review.
Cited by
-
A complex structure of arrestin-2 bound to a G protein-coupled receptor.Cell Res. 2019 Dec;29(12):971-983. doi: 10.1038/s41422-019-0256-2. Epub 2019 Nov 27. Cell Res. 2019. PMID: 31776446 Free PMC article.
-
Quaternary structures of opsin in live cells revealed by FRET spectrometry.Biochem J. 2016 Nov 1;473(21):3819-3836. doi: 10.1042/BCJ20160422. Epub 2016 Sep 13. Biochem J. 2016. PMID: 27623775 Free PMC article.
-
Novel fluorescent GPCR biosensor detects retinal equilibrium binding to opsin and active G protein and arrestin signaling conformations.J Biol Chem. 2020 Dec 18;295(51):17486-17496. doi: 10.1074/jbc.RA120.014631. J Biol Chem. 2020. PMID: 33453993 Free PMC article.
-
Functional map of arrestin-1 at single amino acid resolution.Proc Natl Acad Sci U S A. 2014 Feb 4;111(5):1825-30. doi: 10.1073/pnas.1319402111. Epub 2014 Jan 21. Proc Natl Acad Sci U S A. 2014. PMID: 24449856 Free PMC article.
-
Formation and decay of the arrestin·rhodopsin complex in native disc membranes.J Biol Chem. 2015 May 15;290(20):12919-28. doi: 10.1074/jbc.M114.620898. Epub 2015 Apr 6. J Biol Chem. 2015. PMID: 25847250 Free PMC article.
References
-
- Park J. H., Scheerer P., Hofmann K. P., Choe H. W. & Ernst O. P. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature 454, 183–187 (2008). - PubMed
-
- Scheerer P. et al.. Crystal structure of opsin in its G-protein-interacting conformation. Nature 455, 497–502 (2008). - PubMed
-
- Hofmann K. P. et al.. A G protein-coupled receptor at work: the rhodopsin model. Trends Biochem. Sci. 34, 540–552 (2009). - PubMed
-
- Choe H. W. et al.. Crystal structure of metarhodopsin II. Nature 471, 651–655 (2011). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

