Biochemical analysis of protein SUMOylation
- PMID: 22870855
- PMCID: PMC3477621
- DOI: 10.1002/0471142727.mb1029s99
Biochemical analysis of protein SUMOylation
Abstract
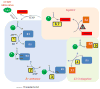
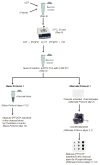
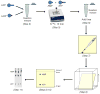
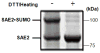
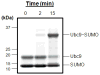
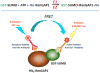
SUMOylation, the covalent attachment of Small Ubiquitin-like MOdifier (SUMO) polypeptides to other proteins, is among the most important post-translational modifications that regulate the functional properties of a large number of proteins. SUMOylation is broadly involved in cellular processes such as gene transcription, hormone response, signal transduction, DNA repair, and nuclear transport. SUMO modification has also been implicated in the pathogenesis of human diseases, such as cancer, neurodegenerative disorders, and viral infection. Attachment of a SUMO protein to another protein is carried out in multiple steps catalyzed by three enzymes. This unit describes and discusses the in vitro biochemical methods used for investigating each step of the SUMOylation process. In addition, a high-throughput screening protocol is included for the identification of inhibitors of SUMOylation.
Figures



Similar articles
-
Preparation of sumoylated substrates for biochemical analysis.Methods Mol Biol. 2009;497:201-10. doi: 10.1007/978-1-59745-566-4_13. Methods Mol Biol. 2009. PMID: 19107419 Review.
-
In vitro assay to determine SUMOylation sites on protein substrates.Nat Protoc. 2016 Feb;11(2):387-97. doi: 10.1038/nprot.2016.023. Epub 2016 Jan 28. Nat Protoc. 2016. PMID: 26820795
-
Identification of a new small ubiquitin-like modifier (SUMO)-interacting motif in the E3 ligase PIASy.J Biol Chem. 2017 Jun 16;292(24):10230-10238. doi: 10.1074/jbc.M117.789982. Epub 2017 Apr 28. J Biol Chem. 2017. PMID: 28455449 Free PMC article.
-
Small-molecule inhibitors targeting small ubiquitin-like modifier pathway for the treatment of cancers and other diseases.Eur J Med Chem. 2022 Apr 5;233:114227. doi: 10.1016/j.ejmech.2022.114227. Epub 2022 Feb 28. Eur J Med Chem. 2022. PMID: 35247754 Review.
-
Assays for investigating deSUMOylation enzymes.Curr Protoc Mol Biol. 2012 Jul;Chapter 10:Unit10.30. doi: 10.1002/0471142727.mb1030s99. Curr Protoc Mol Biol. 2012. PMID: 22870856 Free PMC article.
Cited by
-
Sumoylation of RORγt regulates TH17 differentiation and thymocyte development.Nat Commun. 2018 Nov 19;9(1):4870. doi: 10.1038/s41467-018-07203-z. Nat Commun. 2018. PMID: 30451821 Free PMC article.
-
Sumoylation of histone deacetylase 1 regulates MyoD signaling during myogenesis.Exp Mol Med. 2018 Jan 12;50(1):e427. doi: 10.1038/emm.2017.236. Exp Mol Med. 2018. PMID: 29328071 Free PMC article.
-
Control of regional decidualization in implantation: Role of FoxM1 downstream of Hoxa10 and cyclin D3.Sci Rep. 2015 Sep 9;5:13863. doi: 10.1038/srep13863. Sci Rep. 2015. PMID: 26350477 Free PMC article.
-
Allosteric Inhibition of Ubiquitin-like Modifications by a Class of Inhibitor of SUMO-Activating Enzyme.Cell Chem Biol. 2019 Feb 21;26(2):278-288.e6. doi: 10.1016/j.chembiol.2018.10.026. Epub 2018 Dec 20. Cell Chem Biol. 2019. PMID: 30581133 Free PMC article.
-
An electrophoretic mobility shift assay identifies a mechanistically unique inhibitor of protein sumoylation.Chem Biol. 2013 Apr 18;20(4):604-13. doi: 10.1016/j.chembiol.2013.04.001. Chem Biol. 2013. PMID: 23601649 Free PMC article.
References
-
- Bruzzese FJ, Tsu CA, Ma J, Loke H-K, Wu D, Li Z, Tayber O, Dick LR. Development of a charcoal paper adenosine triphosphate:pyrophosphate exchange assay: kinetic characterization of NEDD8 activating enzyme. Anal Biochem. 2009;394:24–29. - PubMed
-
- Desterro JM, Rodriguez MS, Hay RT. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol Cell. 1998;2:233–239. - PubMed
-
- Desterro JMP, Rodriguez MS, Kemp GD, Hay RT. Identification of the enzyme required for activation of the small ubiquitin-like protein SUMO-1. J Biol Chem. 1999;274:10618–10624. - PubMed
-
- Dhananjayan SC, Ismail A, Nawaz Z. Ubiquitin and control of transcription. In: Mayer RJLR, editor. Essays Biochem, Vol 41: The Ubiquitin-Proteasome System. 2005. pp. 69–80. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

