Non structural protein of avian influenza A (H11N1) virus is a weaker suppressor of immune responses but capable of inducing apoptosis in host cells
- PMID: 22866982
- PMCID: PMC3490754
- DOI: 10.1186/1743-422X-9-149
Non structural protein of avian influenza A (H11N1) virus is a weaker suppressor of immune responses but capable of inducing apoptosis in host cells
Abstract
Background: The Non-Structural (NS1) protein of Influenza A viruses is an extensively studied multifunctional protein which is commonly considered as key viral component to fight against host immune responses. Even though there has been a lot of studies on the involvement of NS1 protein in host immune responses there are still ambiguities regarding its role in apoptosis in infected cells. Interactions of NS1 protein with host factors, role of NS1 protein in regulating cellular responses and apoptosis are quite complicated and further studies are still needed to understand it completely.
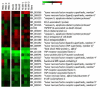
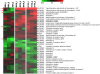
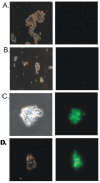
Results: NS1 genes of influenza A/Chicken/India/WBNIV2653/2008 (H5N1) and A/Aquatic bird/India/NIV-17095/2007(H11N1) were cloned and expressed in human embryonic kidney (293T) cells. Microarray based approach to study the host cellular responses to NS1 protein of the two influenza A viruses of different pathogenicity showed significant differences in the host gene expression profile. NS1 protein of H5N1 resulted in suppression of IFN-β mediated innate immune responses, leading to down-regulation of the components of JAK-STAT pathway like STAT1 which further suppressed the expression of pro-inflammatory cytokines like CXCL10 and CCL5. The degree of suppression of host immune genes was found considerable with NS1 protein of H11N1 but was not as prominent as with H5N1-NS1. TUNEL assay analyses were found to be positive in both the NS1 transfected cells indicating both H5N1 as well as H11N1 NS1 proteins were able to induce apoptosis in transfected cells.
Conclusions: We propose that NS1 protein of both H5N1 and H11N1 subtypes of influenza viruses are capable of influencing host immune responses and possess necessary functionality to support apoptosis in host cells. H11N1, a low pathogenic virus without any proven evidence to infect mammals, contains a highly potential NS1 gene which might contribute to greater virus virulence in different gene combinations.
Figures


Similar articles
-
Differences in Type I interferon response in human lung epithelial cells infected by highly pathogenic H5N1 and low pathogenic H11N1 avian influenza viruses.Virus Genes. 2018 Jun;54(3):414-423. doi: 10.1007/s11262-018-1556-1. Epub 2018 Mar 24. Virus Genes. 2018. PMID: 29574656
-
Mammalian Adaptation of an Avian Influenza A Virus Involves Stepwise Changes in NS1.J Virol. 2018 Feb 12;92(5):e01875-17. doi: 10.1128/JVI.01875-17. Print 2018 Mar 1. J Virol. 2018. PMID: 29237841 Free PMC article.
-
Differential Modulation of Innate Immune Responses in Human Primary Cells by Influenza A Viruses Carrying Human or Avian Nonstructural Protein 1.J Virol. 2019 Dec 12;94(1):e00999-19. doi: 10.1128/JVI.00999-19. Print 2019 Dec 12. J Virol. 2019. PMID: 31597767 Free PMC article.
-
Modulation of Innate Immune Responses by the Influenza A NS1 and PA-X Proteins.Viruses. 2018 Dec 12;10(12):708. doi: 10.3390/v10120708. Viruses. 2018. PMID: 30545063 Free PMC article. Review.
-
Research progress on the nonstructural protein 1 (NS1) of influenza a virus.Virulence. 2024 Dec;15(1):2359470. doi: 10.1080/21505594.2024.2359470. Epub 2024 Jun 25. Virulence. 2024. PMID: 38918890 Free PMC article. Review.
Cited by
-
Identification of a Highly Conserved Epitope on Avian Influenza Virus Non-Structural Protein 1 Using a Peptide Microarray.PLoS One. 2016 Mar 3;11(3):e0149868. doi: 10.1371/journal.pone.0149868. eCollection 2016. PLoS One. 2016. PMID: 26938453 Free PMC article.
-
The Contribution of Viral Proteins to the Synergy of Influenza and Bacterial Co-Infection.Viruses. 2022 May 16;14(5):1064. doi: 10.3390/v14051064. Viruses. 2022. PMID: 35632805 Free PMC article. Review.
-
NS1: A Key Protein in the "Game" Between Influenza A Virus and Host in Innate Immunity.Front Cell Infect Microbiol. 2021 Jul 13;11:670177. doi: 10.3389/fcimb.2021.670177. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34327148 Free PMC article. Review.
-
Highly pathogenic avian influenza A virus H5N1 non-structural protein 1 is associated with apoptotic activation of the intrinsic mitochondrial pathway.Exp Ther Med. 2017 Nov;14(5):4041-4046. doi: 10.3892/etm.2017.5056. Epub 2017 Aug 28. Exp Ther Med. 2017. PMID: 29067097 Free PMC article.
-
Human Influenza Virus Infections.Semin Respir Crit Care Med. 2016 Aug;37(4):487-500. doi: 10.1055/s-0036-1584801. Epub 2016 Aug 3. Semin Respir Crit Care Med. 2016. PMID: 27486731 Free PMC article. Review.
References
-
- Palese P, Shaw ML. In: Fields Virology. 5. Knipe DM, Howley PM, editor. Philadelphia: Lippincott Williams and Wilkins; 2008. Orthomyxoviridae: The viruses and their replication; pp. 1647–1689.
-
- Gack MU, Albrecht RA, Urano T, Inn KS, Huang IC, Carnero E, Farzan M, Inoue S, Jung JU, Garcia-Sastre A. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-1. Cell Host Microbe. 2009;5:439–449. doi: 10.1016/j.chom.2009.04.006. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

